Similar analysis was found in Guangchuang Yu bioconductor vignettes page.
library(tidyverse);library(ShortRead);library(goseq);library(GO.db);library("annotate")load GOseq function
# download cDNA info directly from TAIR
At_cdna<-Biostrings::readDNAStringSet("https://www.arabidopsis.org/download_files/Sequences/TAIR10_blastsets/TAIR10_cds_20110103_representative_gene_model_updated")
At_cdna## A DNAStringSet instance of length 27416
## width seq names
## [1] 2016 ATGGTTCAATATAATTTCA...AAAACCGACAGGCGTTGA AT1G50920.1 | Sym...
## [2] 546 ATGACTCGTTTGTTGCCTT...GTTGATTCTGGTACATAG AT1G36960.1 | Sym...
## [3] 1734 ATGGATTCAGAGTCAGAGT...GATTATATTCTCCAATGA AT1G44020.1 | Sym...
## [4] 1059 ATGTCGGTTCCTCCTAGAT...AGAGAGAGTGATAAATAA AT1G15970.1 | Sym...
## [5] 765 ATGGCGAGGGGAGAATCGG...ATGTTGAAAGGTTCTTGA AT1G73440.1 | Sym...
## ... ... ...
## [27412] 2019 GGGTTGAAGTTTAGACCGC...ACTACTAGTCTAGTCTAG ATMG00520.1 | Sym...
## [27413] 303 ATGGATCTTATCAAATATT...AATAGCATTCAAGGTTAA ATMG00650.1 | Sym...
## [27414] 384 ATGCTCCCCGCCGGTTGTT...CGATACTTAACTATATAA ATMG01330.1 | Sym...
## [27415] 573 ATGGATAACCAATTCATTT...CAGCGTAGCGACGGATAA ATMG00070.1 | Sym...
## [27416] 366 ATGGCATCAAAAATCCGCA...CCTTCTGCATACGCATAA ATMG00130.1 | Sym...# GOseq.Atgoslim.BP.list.ORA function
#This function is used to calculate GO ORA results multiple times.
getwd()## [1] "/Volumes/data_personal/Kazu_blog/content/post"load("2018-09-27-over-representation-analysis-4-heatmap-visualization_files/Atgoslim.TAIR.BP.list.Rdata")
head(Atgoslim.TAIR.BP.list)## $`18S RRNA`
## [1] "GO:0006412" "GO:0006412" "GO:0006412"
##
## $`25S RRNA`
## [1] "GO:0043043" "GO:0043043"
##
## $`5.8S RRNA`
## [1] "GO:0043043" "GO:0043043"
##
## $AAN
## [1] "GO:0007623"
##
## $AAR1
## [1] "GO:0006970" "GO:0006970" "GO:0040029"
##
## $AAR2
## [1] "GO:0009845" "GO:0040029"GOseq.Atgoslim.BP.list.ORA<-function(genelist,padjust=0.05,ontology="BP",custom.category.list=Atgoslim.TAIR.BP.list,cdna=At_cdna) { # return GO enrichment table, padjus, padjust=0.05.
bias<-nchar(cdna)
names(bias)<-tibble(AGI=names(cdna)) %>% separate(AGI,into="AGI2",pattern="|",extra="drop") %>% dplyr::select(AGI2) %>% as_vector()
table(duplicated(names(bias)))
#TF<-(names(bias) %in% genelist)*1
TF<-as.integer(names(bias) %in% genelist)
names(TF)<-names(bias)
#print(TF)
pwf<-nullp(TF,bias.data=bias)
#print(pwf$DEgenes)
GO.pval <- goseq(pwf,gene2cat=custom.category.list,use_genes_without_cat=TRUE) # format became different in new goseq version (021111). Does not work (042716)
#head(GO.pval)
if(ontology=="BP") {
GO.pval2<-subset(GO.pval,ontology=="BP")
} else if(ontology=="CC") {
GO.pval2<-subset(GO.pval,ontology=="CC")
} else {
GO.pval2<-subset(GO.pval,ontology=="MF")
}
# calculating padjust by BH
GO.pval2$over_represented_padjust<-p.adjust(GO.pval2$over_represented_pvalue,method="BH")
if(GO.pval2$over_represented_padjust[1]>padjust) return("no enriched GO")
else {
enriched.GO<-GO.pval2[GO.pval2$over_represented_padjust<padjust,]
print("enriched.GO is")
print(enriched.GO)
## write Term and Definition
for(i in 1:dim(enriched.GO)[1]) {
if(is.null(Term(GOTERM[enriched.GO[i,"category"]]))) {next} else {
enriched.GO$Term[i]<-Term(GOTERM[[enriched.GO[i,"category"]]])
enriched.GO$Definition[i]<-Definition(GOTERM[[enriched.GO[i,"category"]]])
}
}
return(enriched.GO)
}
}Importing enrichment result table and have summary table: Table of DEGs (under construction)
# Ding (2018) Cell DEGs
list.files(path=file.path("2018-10-01-differential-expression-analysis-with-public-available-sequencing-data_files","output")) # finding files in different blogs did not work.. Did work this time... why?## [1] "genotypenpr1_genotypenpr1.treatmenttreated.DEGs.int.rWT.rU.csv"
## [2] "genotypenpr4_genotypenpr4.treatmenttreated.DEGs.int.rWT.rU.csv"
## [3] "trt.DEGs.int.rWT.rU.csv"getwd()## [1] "/Volumes/data_personal/Kazu_blog/content/post"DEG.objs<-list.files(path=file.path("2018-09-27-over-representation-analysis-4-heatmap-visualization_files","output"),pattern="\\.csv$") # under construction
DEG.objs## [1] "genotypenpr1_genotypenpr1.treatmenttreated.DEGs.int.rWT.rU.csv"
## [2] "genotypenpr4_genotypenpr4.treatmenttreated.DEGs.int.rWT.rU.csv"
## [3] "trt.DEGs.int.rWT.rU.csv"# read csv file
DEG.list<-lapply(DEG.objs, function(x) read_csv(paste(file.path("2018-09-27-over-representation-analysis-4-heatmap-visualization_files","output"),"/",x,sep="")))
names(DEG.list)<-gsub(".csv","",DEG.objs)
DEG.list## $genotypenpr1_genotypenpr1.treatmenttreated.DEGs.int.rWT.rU
## # A tibble: 6,694 x 10
## genes logFC.genotypen… logFC.genotypen… logCPM LR PValue FDR
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 AT2G… -4.20 -0.720 6.69 1300. 5.30e-283 1.78e-278
## 2 AT5G… 3.26 0.530 4.76 1000. 5.96e-218 1.00e-213
## 3 AT1G… -2.02 -0.475 7.19 629. 2.69e-137 3.01e-133
## 4 AT1G… -2.26 -0.511 7.51 620. 2.83e-135 2.38e-131
## 5 AT3G… -3.16 0.888 5.33 618. 5.42e-135 3.64e-131
## 6 AT5G… -0.212 -6.72 3.89 607. 1.67e-132 9.35e-129
## 7 AT2G… 2.22 1.95 9.21 572. 7.80e-125 3.74e-121
## 8 AT5G… 1.82 1.83 9.40 570. 1.54e-124 6.46e-121
## 9 AT1G… -3.65 -1.52 3.37 565. 1.92e-123 7.17e-120
## 10 AT5G… 0.000149 -7.96 3.77 546. 2.42e-119 8.12e-116
## # ... with 6,684 more rows, and 3 more variables: AGI <chr>,
## # symbol2 <chr>, full_name <chr>
##
## $genotypenpr4_genotypenpr4.treatmenttreated.DEGs.int.rWT.rU
## # A tibble: 846 x 10
## genes logFC.genotypen… logFC.genotypen… logCPM LR PValue FDR
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 AT5G… 3.04 0.628 4.76 904. 5.00e-197 1.68e-192
## 2 AT3G… -5.45 -0.183 1.98 459. 1.80e-100 3.03e- 96
## 3 AT5G… -3.30 0.354 3.10 444. 4.51e- 97 5.05e- 93
## 4 AT1G… -7.40 -3.70 2.00 432. 1.32e- 94 1.11e- 90
## 5 AT3G… -2.97 -0.301 3.36 372. 1.83e- 81 1.23e- 77
## 6 AT1G… -2.50 0.891 6.30 288. 2.54e- 63 1.42e- 59
## 7 AT2G… -1.42 -0.703 6.69 277. 7.63e- 61 3.66e- 57
## 8 AT5G… -3.04 0.434 4.19 211. 1.75e- 46 7.36e- 43
## 9 AT2G… -1.10 -0.237 5.87 209. 5.09e- 46 1.90e- 42
## 10 AT5G… -1.84 -0.489 2.78 187. 2.68e- 41 9.01e- 38
## # ... with 836 more rows, and 3 more variables: AGI <chr>, symbol2 <chr>,
## # full_name <chr>
##
## $trt.DEGs.int.rWT.rU
## # A tibble: 7,811 x 9
## genes logFC logCPM LR PValue FDR AGI symbol2 full_name
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <chr>
## 1 AT5G1… 8.57 3.89 675. 7.55e-149 2.54e-144 AT5G… AT-HSP… heat shock…
## 2 AT1G0… 4.00 4.60 592. 7.94e-131 1.33e-126 AT1G… <NA> <NA>
## 3 AT5G2… 6.16 4.19 588. 5.77e-130 6.46e-126 AT5G… ATWRKY… ARABIDOPSI…
## 4 AT4G2… 3.85 5.39 586. 1.92e-129 1.59e-125 AT4G… CRK14 cysteine-r…
## 5 AT5G1… 9.64 3.77 586. 2.37e-129 1.59e-125 AT5G… HSP17.… 17.6 kDa c…
## 6 AT3G2… 4.92 3.36 546. 9.68e-121 5.42e-117 AT3G… <NA> <NA>
## 7 AT4G2… 3.34 6.70 539. 2.78e-119 1.33e-115 AT4G… SGT1A <NA>
## 8 AT5G2… 3.82 6.70 535. 2.42e-118 1.02e-114 AT5G… AtDMR6… NA;DOWNY M…
## 9 AT5G0… 4.14 7.14 523. 1.07e-115 3.98e-112 AT5G… AtHsp7… NA;NA
## 10 AT1G2… 2.87 7.19 511. 3.43e-113 1.15e-109 AT1G… AtWAK1… NA;NA;cell…
## # ... with 7,801 more rows# GOseq.Atgoslim.BP.list.ORA(genelist=genes.shade1h.up$AGI)GO.ORA for each DEG list (loop)
# single coefficient
names(DEG.list)## [1] "genotypenpr1_genotypenpr1.treatmenttreated.DEGs.int.rWT.rU"
## [2] "genotypenpr4_genotypenpr4.treatmenttreated.DEGs.int.rWT.rU"
## [3] "trt.DEGs.int.rWT.rU"for(n in 3:3) {
library(dplyr)
#genelist.up<-base::get(paste("../output/",DEG.objs.V1.5annotation.unique[n],sep="")) %>% rownames_to_column() %>% dplyr::filter(logFC>0) # does not work
genelist.up<-DEG.list[[n]] %>% dplyr::filter(logFC>0)
genelist.down<-DEG.list[[n]] %>% dplyr::filter(logFC<0)
GO.ORA.temp.up<-GOseq.Atgoslim.BP.list.ORA(str_remove(as_vector(genelist.up[,"genes"]),"\\.[[:digit:]]+"),custom.category.list=Atgoslim.TAIR.BP.list)
GO.ORA.temp.down<-GOseq.Atgoslim.BP.list.ORA(str_remove(as_vector(genelist.down[,"genes"]),"\\.[[:digit:]]+"),custom.category.list=Atgoslim.TAIR.BP.list)
# handling "no enriched GO"
# genelist.names<-c("GO.ORA.temp.up_down","GO.ORA.temp.down_up") # test
x<-list(GO.ORA.temp.up=GO.ORA.temp.up,
GO.ORA.temp.down=GO.ORA.temp.down) # list
x<-x[!x=="no enriched GO"] # reove "no enriched GO" result
## add sample info and FC info and save GO.ORA result
for (i in 1:length(x)) {
GO.ORA.result<-x[[i]] %>% mutate(FC = gsub("(GO.ORA.temp.)(.)","\\2",names(x)[i]),sample=DEG.objs[n])
save(GO.ORA.result,file=file.path("2018-09-27-over-representation-analysis-4-heatmap-visualization_files","output",paste(gsub(".csv","",DEG.objs[n]),gsub("(GO.ORA.temp.)(.)","\\2",names(x)[i]),"enrich.Rdata",sep=".")))
rm(GO.ORA.result)
}
}## Warning in pcls(G): initial point very close to some inequality constraints## [1] "enriched.GO is"
## category over_represented_pvalue under_represented_pvalue
## 643 GO:0006952 1.127194e-34 1.0000000
## 624 GO:0006886 7.258753e-33 1.0000000
## 2259 GO:0042742 6.626442e-21 1.0000000
## 1225 GO:0010200 3.858737e-19 1.0000000
## 430 GO:0006468 3.007738e-17 1.0000000
## 626 GO:0006888 2.027405e-16 1.0000000
## 863 GO:0009408 8.132071e-16 1.0000000
## 1453 GO:0015031 1.469037e-15 1.0000000
## 1587 GO:0016192 1.650887e-13 1.0000000
## 1949 GO:0032482 4.969469e-13 1.0000000
## 628 GO:0006891 4.154328e-12 1.0000000
## 901 GO:0009611 7.570706e-12 1.0000000
## 984 GO:0009751 6.974160e-11 1.0000000
## 910 GO:0009626 1.312138e-10 1.0000000
## 2097 GO:0034605 1.472659e-10 1.0000000
## 905 GO:0009617 3.375985e-10 1.0000000
## 3272 GO:0080167 5.223324e-10 1.0000000
## 191 GO:0002237 8.378504e-10 1.0000000
## 717 GO:0007165 8.605229e-10 1.0000000
## 932 GO:0009651 1.039053e-09 1.0000000
## 2245 GO:0042542 4.435876e-09 1.0000000
## 627 GO:0006890 6.596583e-09 1.0000000
## 1014 GO:0009816 1.376798e-08 1.0000000
## 906 GO:0009620 1.712451e-08 1.0000000
## 851 GO:0009306 6.005123e-08 1.0000000
## 1837 GO:0030968 6.276610e-08 1.0000000
## 972 GO:0009737 7.520204e-08 1.0000000
## 675 GO:0007030 8.265301e-08 1.0000000
## 718 GO:0007166 9.044827e-08 1.0000000
## 2803 GO:0051259 1.299325e-07 1.0000000
## 973 GO:0009738 4.810860e-07 1.0000000
## 2871 GO:0051707 2.090818e-06 0.9999995
## 2591 GO:0046777 2.236596e-06 0.9999991
## 654 GO:0006986 2.438230e-06 0.9999998
## 3444 GO:1900150 6.399834e-06 0.9999993
## 3056 GO:0070973 8.359768e-06 0.9999996
## 868 GO:0009414 8.391875e-06 0.9999959
## 2693 GO:0048544 9.648966e-06 0.9999983
## 2450 GO:0045332 1.053543e-05 0.9999995
## 3191 GO:0072583 1.057949e-05 0.9999989
## 1015 GO:0009817 1.361175e-05 0.9999968
## 192 GO:0002238 1.484186e-05 0.9999987
## 1220 GO:0010193 1.562249e-05 0.9999972
## 1198 GO:0010150 1.702895e-05 0.9999937
## 2100 GO:0034620 1.733070e-05 0.9999979
## 429 GO:0006465 1.760197e-05 0.9999985
## 911 GO:0009627 1.774452e-05 0.9999956
## 448 GO:0006499 1.973024e-05 0.9999954
## 1870 GO:0031204 2.235051e-05 1.0000000
## 635 GO:0006904 2.831111e-05 0.9999936
## 1044 GO:0009863 3.189073e-05 0.9999955
## 862 GO:0009407 3.201744e-05 0.9999926
## 511 GO:0006616 3.545040e-05 0.9999976
## 426 GO:0006457 3.979662e-05 0.9999802
## 756 GO:0008219 4.780393e-05 0.9999880
## 925 GO:0009644 4.998914e-05 0.9999851
## 3254 GO:0080142 6.226918e-05 0.9999959
## 625 GO:0006887 6.885707e-05 0.9999810
## 1647 GO:0018105 8.127620e-05 0.9999718
## 903 GO:0009615 8.144403e-05 0.9999805
## 563 GO:0006749 8.501484e-05 0.9999771
## 3185 GO:0072334 8.701453e-05 0.9999978
## 59 GO:0000302 8.708749e-05 0.9999779
## 636 GO:0006906 9.447106e-05 0.9999786
## 2760 GO:0050829 9.815163e-05 0.9999858
## 2158 GO:0035556 9.845166e-05 0.9999520
## 1181 GO:0010112 1.187545e-04 0.9999892
## 651 GO:0006979 1.230762e-04 0.9999318
## 2885 GO:0051865 1.245660e-04 0.9999864
## 3107 GO:0071323 1.510425e-04 1.0000000
## 1043 GO:0009862 1.527420e-04 0.9999828
## 2626 GO:0048194 1.528899e-04 0.9999908
## 2202 GO:0042026 1.836214e-04 0.9999643
## 864 GO:0009409 2.019371e-04 0.9998822
## 679 GO:0007034 2.056534e-04 0.9999640
## 603 GO:0006839 2.267988e-04 0.9999409
## 2974 GO:0061025 2.845410e-04 0.9999373
## 1595 GO:0016310 2.987777e-04 0.9998090
## 1245 GO:0010224 3.068315e-04 0.9998946
## 2778 GO:0051085 3.451232e-04 0.9999110
## 2642 GO:0048278 3.840641e-04 0.9999261
## 1750 GO:0019722 4.127109e-04 0.9998973
## 2584 GO:0046686 4.432858e-04 0.9997292
## 38 GO:0000187 4.616590e-04 0.9999579
## 615 GO:0006874 4.684844e-04 0.9998825
## 2963 GO:0060866 5.252189e-04 0.9999699
## 2761 GO:0050832 5.600444e-04 0.9996520
## 3704 GO:2000022 5.601390e-04 0.9998749
## 2329 GO:0043207 5.966944e-04 1.0000000
## 1013 GO:0009814 6.379255e-04 0.9998552
## 2436 GO:0045087 6.607522e-04 0.9997520
## 504 GO:0006605 7.120692e-04 0.9999344
## 2247 GO:0042546 7.400827e-04 0.9997560
## 2901 GO:0052544 7.413370e-04 0.9998885
## 2458 GO:0045489 7.769621e-04 0.9998080
## 3457 GO:1900425 8.788298e-04 0.9999016
## 1875 GO:0031347 9.014185e-04 0.9996995
## 1777 GO:0023014 1.012589e-03 0.9996473
## 1189 GO:0010120 1.062200e-03 0.9998769
## 3458 GO:1900426 1.073764e-03 0.9998157
## 1853 GO:0031098 1.087543e-03 0.9996323
## 440 GO:0006486 1.167370e-03 0.9994398
## 2207 GO:0042147 1.196837e-03 0.9997222
## 1876 GO:0031348 1.370772e-03 0.9996884
## 189 GO:0002229 1.383542e-03 0.9996077
## numDEInCat numInCat
## 643 228 620
## 624 112 237
## 2259 113 288
## 1225 64 136
## 430 235 774
## 626 47 93
## 863 66 157
## 1453 87 253
## 1587 54 132
## 1949 31 62
## 628 29 48
## 901 72 204
## 984 58 162
## 910 34 67
## 2097 30 58
## 905 45 111
## 3272 49 134
## 191 19 27
## 717 102 351
## 932 133 522
## 2245 30 68
## 627 21 36
## 1014 23 40
## 906 34 78
## 851 18 30
## 1837 13 17
## 972 105 409
## 675 22 42
## 718 24 46
## 2803 11 17
## 973 61 211
## 2871 23 56
## 2591 67 210
## 654 12 17
## 3444 12 19
## 3056 8 11
## 868 73 293
## 2693 17 31
## 2450 10 12
## 3191 12 18
## 1015 20 46
## 192 10 15
## 1220 15 31
## 1198 34 104
## 2100 12 20
## 429 9 15
## 911 20 51
## 448 19 45
## 1870 5 5
## 635 18 39
## 1044 12 22
## 862 17 46
## 511 8 12
## 426 58 231
## 756 19 44
## 925 22 64
## 3254 8 11
## 625 20 50
## 1647 27 74
## 903 17 39
## 563 18 52
## 3185 6 7
## 59 17 46
## 636 15 38
## 2760 11 20
## 2158 50 173
## 1181 8 13
## 651 67 291
## 2885 9 15
## 3107 5 5
## 1043 9 15
## 2626 8 10
## 2202 13 26
## 864 74 329
## 679 11 24
## 603 16 41
## 2974 13 31
## 1595 101 432
## 1245 22 65
## 2778 15 38
## 2642 11 25
## 1750 13 42
## 2584 79 341
## 38 7 11
## 615 15 32
## 2963 6 8
## 2761 76 477
## 3704 12 29
## 2329 3 3
## 1013 12 29
## 2436 23 69
## 504 6 11
## 2247 18 55
## 2901 9 16
## 2458 13 31
## 3457 7 12
## 1875 18 53
## 1777 19 56
## 1189 7 12
## 3458 9 18
## 1853 18 52
## 440 34 119
## 2207 11 28
## 1876 11 25
## 189 14 35
## term
## 643 defense response
## 624 intracellular protein transport
## 2259 defense response to bacterium
## 1225 response to chitin
## 430 protein phosphorylation
## 626 ER to Golgi vesicle-mediated transport
## 863 response to heat
## 1453 protein transport
## 1587 vesicle-mediated transport
## 1949 Rab protein signal transduction
## 628 intra-Golgi vesicle-mediated transport
## 901 response to wounding
## 984 response to salicylic acid
## 910 plant-type hypersensitive response
## 2097 cellular response to heat
## 905 response to bacterium
## 3272 response to karrikin
## 191 response to molecule of bacterial origin
## 717 signal transduction
## 932 response to salt stress
## 2245 response to hydrogen peroxide
## 627 retrograde vesicle-mediated transport, Golgi to ER
## 1014 defense response to bacterium, incompatible interaction
## 906 response to fungus
## 851 protein secretion
## 1837 endoplasmic reticulum unfolded protein response
## 972 response to abscisic acid
## 675 Golgi organization
## 718 cell surface receptor signaling pathway
## 2803 protein complex oligomerization
## 973 abscisic acid-activated signaling pathway
## 2871 response to other organism
## 2591 protein autophosphorylation
## 654 response to unfolded protein
## 3444 regulation of defense response to fungus
## 3056 protein localization to endoplasmic reticulum exit site
## 868 response to water deprivation
## 2693 recognition of pollen
## 2450 phospholipid translocation
## 3191 clathrin-dependent endocytosis
## 1015 defense response to fungus, incompatible interaction
## 192 response to molecule of fungal origin
## 1220 response to ozone
## 1198 leaf senescence
## 2100 cellular response to unfolded protein
## 429 signal peptide processing
## 911 systemic acquired resistance
## 448 N-terminal protein myristoylation
## 1870 posttranslational protein targeting to membrane, translocation
## 635 vesicle docking involved in exocytosis
## 1044 salicylic acid mediated signaling pathway
## 862 toxin catabolic process
## 511 SRP-dependent cotranslational protein targeting to membrane, translocation
## 426 protein folding
## 756 cell death
## 925 response to high light intensity
## 3254 regulation of salicylic acid biosynthetic process
## 625 exocytosis
## 1647 peptidyl-serine phosphorylation
## 903 response to virus
## 563 glutathione metabolic process
## 3185 UDP-galactose transmembrane transport
## 59 response to reactive oxygen species
## 636 vesicle fusion
## 2760 defense response to Gram-negative bacterium
## 2158 intracellular signal transduction
## 1181 regulation of systemic acquired resistance
## 651 response to oxidative stress
## 2885 protein autoubiquitination
## 3107 cellular response to chitin
## 1043 systemic acquired resistance, salicylic acid mediated signaling pathway
## 2626 Golgi vesicle budding
## 2202 protein refolding
## 864 response to cold
## 679 vacuolar transport
## 603 mitochondrial transport
## 2974 membrane fusion
## 1595 phosphorylation
## 1245 response to UV-B
## 2778 chaperone cofactor-dependent protein refolding
## 2642 vesicle docking
## 1750 calcium-mediated signaling
## 2584 response to cadmium ion
## 38 activation of MAPK activity
## 615 cellular calcium ion homeostasis
## 2963 leaf abscission
## 2761 defense response to fungus
## 3704 regulation of jasmonic acid mediated signaling pathway
## 2329 response to external biotic stimulus
## 1013 defense response, incompatible interaction
## 2436 innate immune response
## 504 protein targeting
## 2247 cell wall biogenesis
## 2901 defense response by callose deposition in cell wall
## 2458 pectin biosynthetic process
## 3457 negative regulation of defense response to bacterium
## 1875 regulation of defense response
## 1777 signal transduction by protein phosphorylation
## 1189 camalexin biosynthetic process
## 3458 positive regulation of defense response to bacterium
## 1853 stress-activated protein kinase signaling cascade
## 440 protein glycosylation
## 2207 retrograde transport, endosome to Golgi
## 1876 negative regulation of defense response
## 189 defense response to oomycetes
## ontology over_represented_padjust
## 643 BP 4.265301e-31
## 624 BP 1.373356e-29
## 2259 BP 8.358152e-18
## 1225 BP 3.650365e-16
## 430 BP 2.276256e-14
## 626 BP 1.278617e-13
## 863 BP 4.395965e-13
## 1453 BP 6.948544e-13
## 1587 BP 6.941063e-11
## 1949 BP 1.880447e-10
## 628 BP 1.429089e-09
## 901 BP 2.387296e-09
## 984 BP 2.030017e-08
## 910 BP 3.546523e-08
## 2097 BP 3.715029e-08
## 905 BP 7.984205e-08
## 3272 BP 1.162651e-07
## 191 BP 1.713799e-07
## 717 BP 1.713799e-07
## 932 BP 1.965887e-07
## 2245 BP 7.993026e-07
## 627 BP 1.134612e-06
## 1014 BP 2.265132e-06
## 906 BP 2.699965e-06
## 851 BP 9.089355e-06
## 1837 BP 9.134882e-06
## 972 BP 1.053943e-05
## 675 BP 1.116996e-05
## 718 BP 1.180194e-05
## 2803 BP 1.638882e-05
## 973 BP 5.872352e-05
## 2871 BP 2.472392e-04
## 2591 BP 2.564630e-04
## 654 BP 2.713606e-04
## 3444 BP 6.919135e-04
## 3056 BP 8.582393e-04
## 868 BP 8.582393e-04
## 2693 BP 9.608339e-04
## 2450 BP 1.000819e-03
## 3191 BP 1.000819e-03
## 1015 BP 1.256265e-03
## 192 BP 1.337181e-03
## 1220 BP 1.374779e-03
## 1198 BP 1.428623e-03
## 2100 BP 1.428623e-03
## 429 BP 1.428623e-03
## 911 BP 1.428623e-03
## 448 BP 1.555400e-03
## 1870 BP 1.726006e-03
## 635 BP 2.142585e-03
## 1044 BP 2.329885e-03
## 862 BP 2.329885e-03
## 511 BP 2.531025e-03
## 426 BP 2.788711e-03
## 756 BP 3.288910e-03
## 925 BP 3.377838e-03
## 3254 BP 4.133800e-03
## 625 BP 4.492330e-03
## 1647 BP 5.136403e-03
## 903 BP 5.136403e-03
## 563 BP 5.230779e-03
## 3185 BP 5.230779e-03
## 59 BP 5.230779e-03
## 636 BP 5.585601e-03
## 2760 BP 5.644562e-03
## 2158 BP 5.644562e-03
## 1181 BP 6.706971e-03
## 651 BP 6.831271e-03
## 2885 BP 6.831271e-03
## 3107 BP 8.035214e-03
## 1043 BP 8.035214e-03
## 2626 BP 8.035214e-03
## 2202 BP 9.518129e-03
## 864 BP 1.032608e-02
## 679 BP 1.037590e-02
## 603 BP 1.129219e-02
## 2974 BP 1.398316e-02
## 1595 BP 1.449455e-02
## 1245 BP 1.469684e-02
## 2778 BP 1.632433e-02
## 2642 BP 1.794196e-02
## 1750 BP 1.904510e-02
## 2584 BP 2.020956e-02
## 38 BP 2.079664e-02
## 615 BP 2.085582e-02
## 2963 BP 2.310963e-02
## 2761 BP 2.408598e-02
## 3704 BP 2.408598e-02
## 2329 BP 2.536957e-02
## 1013 BP 2.682122e-02
## 2436 BP 2.747567e-02
## 504 BP 2.928772e-02
## 2247 BP 2.984276e-02
## 2901 BP 2.984276e-02
## 2458 BP 3.094763e-02
## 3457 BP 3.464054e-02
## 1875 BP 3.516462e-02
## 1777 BP 3.909835e-02
## 1189 BP 4.059965e-02
## 3458 BP 4.063123e-02
## 1853 BP 4.074516e-02
## 440 BP 4.330714e-02
## 2207 BP 4.396922e-02
## 1876 BP 4.986022e-02
## 189 BP 4.986022e-02## Warning in pcls(G): initial point very close to some inequality constraints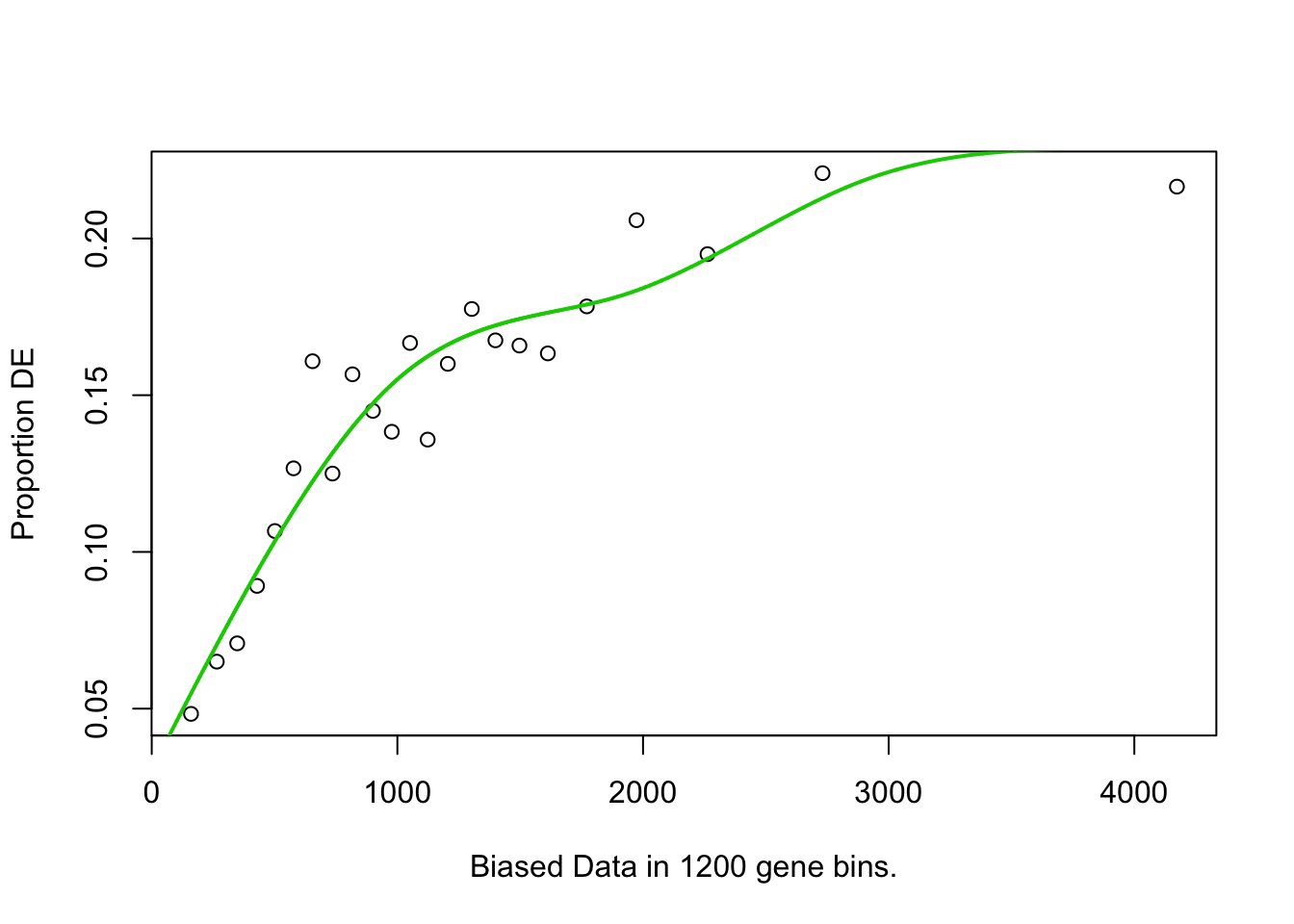
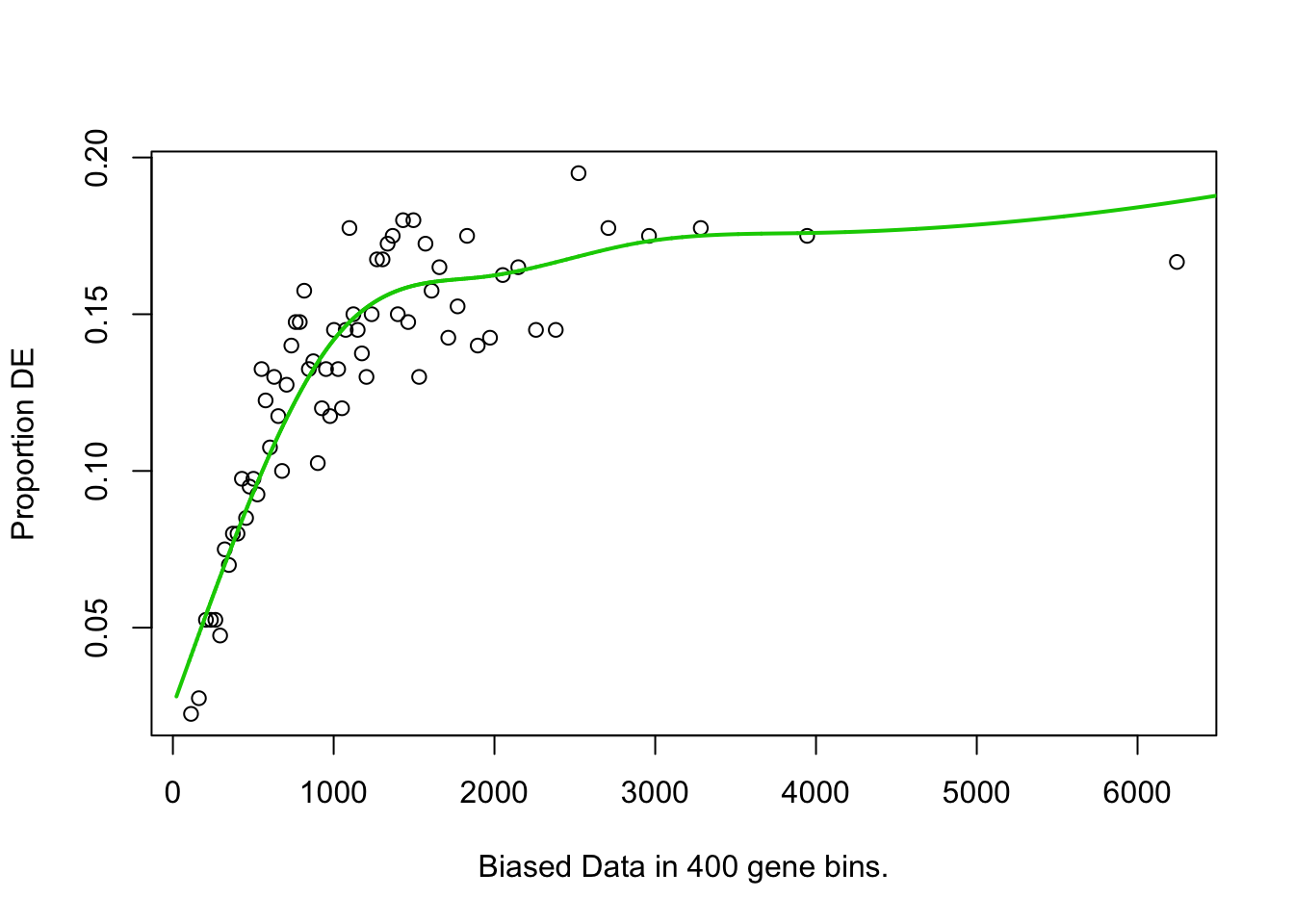
## [1] "enriched.GO is"
## category over_represented_pvalue under_represented_pvalue
## 936 GO:0009658 1.421084e-23 1.0000000
## 1542 GO:0015979 4.134451e-23 1.0000000
## 870 GO:0009416 4.934159e-14 1.0000000
## 990 GO:0009768 1.601237e-13 1.0000000
## 3480 GO:1901259 2.967639e-12 1.0000000
## 1547 GO:0015995 6.409151e-10 1.0000000
## 993 GO:0009773 1.723458e-08 1.0000000
## 745 GO:0007623 4.511876e-08 1.0000000
## 926 GO:0009645 6.696258e-08 1.0000000
## 2278 GO:0042793 4.800680e-07 1.0000000
## 918 GO:0009637 5.268320e-07 0.9999999
## 1118 GO:0010027 1.246089e-06 0.9999997
## 1183 GO:0010114 3.999797e-06 0.9999990
## 864 GO:0009409 4.663120e-06 0.9999978
## 2426 GO:0045036 4.748080e-06 0.9999995
## 1270 GO:0010258 1.091101e-05 1.0000000
## 1678 GO:0019252 2.555344e-05 0.9999961
## 974 GO:0009739 4.714442e-05 0.9999824
## 989 GO:0009767 4.782966e-05 0.9999938
## 1232 GO:0010207 5.485926e-05 0.9999922
## 927 GO:0009646 6.023482e-05 0.9999870
## 215 GO:0005983 6.084207e-05 0.9999930
## 3141 GO:0071482 6.262228e-05 0.9999953
## 935 GO:0009657 6.663415e-05 0.9999950
## 3464 GO:1900865 7.848116e-05 0.9999939
## 1575 GO:0016120 8.238966e-05 0.9999979
## 1111 GO:0010020 9.437563e-05 0.9999856
## 925 GO:0009644 9.468408e-05 0.9999717
## 1257 GO:0010239 9.809432e-05 0.9999974
## 67 GO:0000373 1.030097e-04 0.9999867
## 2427 GO:0045037 1.547592e-04 0.9999781
## 991 GO:0009769 2.165699e-04 0.9999855
## 141 GO:0000967 2.272397e-04 0.9999943
## 1882 GO:0031425 2.569678e-04 0.9999737
## 668 GO:0007018 2.652535e-04 0.9999108
## 3465 GO:1900871 3.085897e-04 1.0000000
## 364 GO:0006353 3.581310e-04 0.9999688
## 1239 GO:0010218 4.028751e-04 0.9998846
## 1221 GO:0010196 4.447964e-04 0.9999657
## numDEInCat numInCat
## 936 76 158
## 1542 63 137
## 870 73 210
## 990 19 24
## 3480 15 16
## 1547 22 37
## 993 12 17
## 745 36 100
## 926 9 10
## 2278 9 10
## 918 24 58
## 1118 21 49
## 1183 22 59
## 864 74 329
## 2426 12 20
## 1270 5 5
## 1678 13 25
## 974 30 112
## 989 10 21
## 1232 11 22
## 927 15 37
## 215 10 17
## 3141 8 12
## 935 8 12
## 3464 8 12
## 1575 6 7
## 1111 11 22
## 925 20 64
## 1257 6 7
## 67 10 18
## 2427 10 19
## 991 6 10
## 141 5 6
## 1882 8 13
## 668 22 62
## 3465 4 4
## 364 7 11
## 1239 16 48
## 1221 6 10
## term ontology
## 936 chloroplast organization BP
## 1542 photosynthesis BP
## 870 response to light stimulus BP
## 990 photosynthesis, light harvesting in photosystem I BP
## 3480 chloroplast rRNA processing BP
## 1547 chlorophyll biosynthetic process BP
## 993 photosynthetic electron transport in photosystem I BP
## 745 circadian rhythm BP
## 926 response to low light intensity stimulus BP
## 2278 plastid transcription BP
## 918 response to blue light BP
## 1118 thylakoid membrane organization BP
## 1183 response to red light BP
## 864 response to cold BP
## 2426 protein targeting to chloroplast BP
## 1270 NADH dehydrogenase complex (plastoquinone) assembly BP
## 1678 starch biosynthetic process BP
## 974 response to gibberellin BP
## 989 photosynthetic electron transport chain BP
## 1232 photosystem II assembly BP
## 927 response to absence of light BP
## 215 starch catabolic process BP
## 3141 cellular response to light stimulus BP
## 935 plastid organization BP
## 3464 chloroplast RNA modification BP
## 1575 carotene biosynthetic process BP
## 1111 chloroplast fission BP
## 925 response to high light intensity BP
## 1257 chloroplast mRNA processing BP
## 67 Group II intron splicing BP
## 2427 protein import into chloroplast stroma BP
## 991 photosynthesis, light harvesting in photosystem II BP
## 141 rRNA 5'-end processing BP
## 1882 chloroplast RNA processing BP
## 668 microtubule-based movement BP
## 3465 chloroplast mRNA modification BP
## 364 DNA-templated transcription, termination BP
## 1239 response to far red light BP
## 1221 nonphotochemical quenching BP
## over_represented_padjust
## 936 5.377382e-20
## 1542 7.822381e-20
## 870 6.223620e-11
## 990 1.514770e-10
## 3480 2.245909e-09
## 1547 4.042038e-07
## 993 9.316524e-06
## 745 2.134117e-05
## 926 2.815404e-05
## 2278 1.812302e-04
## 918 1.812302e-04
## 1118 3.929334e-04
## 1183 1.164249e-03
## 864 1.197782e-03
## 2426 1.197782e-03
## 1270 2.580453e-03
## 1678 5.687894e-03
## 974 9.525655e-03
## 989 9.525655e-03
## 1232 1.030273e-02
## 927 1.030273e-02
## 215 1.030273e-02
## 3141 1.030273e-02
## 935 1.050598e-02
## 3464 1.187891e-02
## 1575 1.199086e-02
## 1111 1.279588e-02
## 925 1.279588e-02
## 1257 1.279962e-02
## 67 1.299295e-02
## 2427 1.889061e-02
## 991 2.560939e-02
## 141 2.605682e-02
## 1882 2.859900e-02
## 668 2.867769e-02
## 3465 3.243620e-02
## 364 3.662615e-02
## 1239 4.011788e-02
## 1221 4.315666e-02do separate analysis for double coefficient type
for(n in 1:2) {
library(dplyr)
#genelist.up<-base::get(paste("../output/",DEG.objs.V1.5annotation.unique[n],sep="")) %>% rownames_to_column() %>% dplyr::filter(logFC>0) # does not work
genelist.all<-DEG.list[[n]]
# DEG.list[[n]] %>% dplyr::select("logFC.genotypenpr1.1")
# DEG.list[[n]] %>% dplyr::select(logFC.genotypenpr1.1)
#
# DEG.list[[n]] %>% dplyr::select(names(DEG.list[[n]])[2]) %>% filter(.>0)
# DEG.list[[n]] %>% dplyr::filter(names(DEG.list[[n]])[2]>0)
# DEG.list[[n]] %>% dplyr::filter(logFC.genotypenpr1.1>0) # does work
# DEG.list[[n]] %>% dplyr::filter("logFC.genotypenpr1.1">0)
# DEG.list[[n]] %>% dplyr::filter(rlang::UQ("logFC.genotypenpr1.1")>0)
# DEG.list[[n]] %>% dplyr::filter(!!"logFC.genotypenpr1.1">0)
# DEG.list[[n]] %>% dplyr::filter(!!names(DEG.list[[n]])[2]>0)
# DEG.list[[n]] %>% dplyr::filter((!!names(DEG.list[[n]])[2])==3.26)
# DEG.list[[n]] %>% dplyr::filter((!!names(DEG.list[[n]])[2])<0)
# DEG.list[[n]] %>% dplyr::filter((UQ(as.name(names(DEG.list[[n]])[2])))>0) # works! See https://stackoverflow.com/questions/27197617/filter-data-frame-by-character-column-name-in-dplyr
# DEG.list[[n]] %>% dplyr::filter((as.name(names(DEG.list[[n]])[2]))>0) # does not work
genelist.upup<-DEG.list[[n]] %>% dplyr::filter((UQ(as.name(names(DEG.list[[n]])[2])))>0) %>% dplyr::filter((UQ(as.name(names(DEG.list[[n]])[3])))>0)
genelist.updown<-DEG.list[[n]] %>% dplyr::filter((UQ(as.name(names(DEG.list[[n]])[2])))>0) %>% dplyr::filter((UQ(as.name(names(DEG.list[[n]])[3])))<0)
genelist.downup<-DEG.list[[n]] %>% dplyr::filter((UQ(as.name(names(DEG.list[[n]])[2])))<0) %>% dplyr::filter((UQ(as.name(names(DEG.list[[n]])[3])))>0)
genelist.downdown<- DEG.list[[n]] %>% dplyr::filter((UQ(as.name(names(DEG.list[[n]])[2])))<0) %>% dplyr::filter((UQ(as.name(names(DEG.list[[n]])[3])))<0)
GO.ORA.temp.all<-GOseq.Atgoslim.BP.list.ORA(str_remove(as_vector(genelist.all[,"genes"]),"\\.[[:digit:]]+"),custom.category.list=Atgoslim.TAIR.BP.list)
GO.ORA.temp.up_up<-GOseq.Atgoslim.BP.list.ORA(str_remove(as_vector(genelist.upup[,"genes"]),"\\.[[:digit:]]+"),custom.category.list=Atgoslim.TAIR.BP.list)
GO.ORA.temp.up_down<-GOseq.Atgoslim.BP.list.ORA(str_remove(as_vector(genelist.updown[,"genes"]),"\\.[[:digit:]]+"),custom.category.list=Atgoslim.TAIR.BP.list)
GO.ORA.temp.down_up<-GOseq.Atgoslim.BP.list.ORA(str_remove(as_vector(genelist.downup[,"genes"]),"\\.[[:digit:]]+"),custom.category.list=Atgoslim.TAIR.BP.list)
GO.ORA.temp.down_down<-GOseq.Atgoslim.BP.list.ORA(str_remove(as_vector(genelist.downdown[,"genes"]),"\\.[[:digit:]]+"),custom.category.list=Atgoslim.TAIR.BP.list)
# handling "no enriched GO"
# genelist.names<-c("GO.ORA.temp.up_down","GO.ORA.temp.down_up") # test
x<-list(GO.ORA.temp.all=GO.ORA.temp.all,
GO.ORA.temp.up_up=GO.ORA.temp.up_up,
GO.ORA.temp.up_down=GO.ORA.temp.up_down,
GO.ORA.temp.down_up=GO.ORA.temp.down_up,
GO.ORA.temp.down_down=GO.ORA.temp.down_down) # list
print(x[x=="no enriched GO"])
x<-x[!x=="no enriched GO"] # reove "no enriched GO" result
## add sample info and FC info and save GO.ORA result
for (i in 1:length(x)) {
GO.ORA.result<-x[[i]] %>% mutate(FC = gsub("(GO.ORA.temp.)(.)","\\2",names(x)[i]),sample=DEG.objs[n])
save(GO.ORA.result,file=file.path("2018-09-27-over-representation-analysis-4-heatmap-visualization_files","output",paste(gsub(".csv","",DEG.objs[n]),gsub("(GO.ORA.temp.)(.)","\\2",names(x)[i]),"enrich.Rdata",sep=".")))
rm(GO.ORA.result)
}
}read GOseq result table
eGOseqs<-list.files(pattern="enrich.Rdata",path=file.path("2018-09-27-over-representation-analysis-4-heatmap-visualization_files","output"))
eGOseqs.list2<-sapply(file.path("2018-09-27-over-representation-analysis-4-heatmap-visualization_files","output",eGOseqs),function(x) mget(load(x))) # mget will return the value of the object(or objects) in a list. see https://stackoverflow.com/questions/29398630/load-data-frames-into-list
#names(eGOseqs.list2)
eGOseqs.list2.summary<-do.call("rbind",eGOseqs.list2)
#head(eGOseqs.list2.summary) # make sure those are file names
rownames(eGOseqs.list2.summary)<-1:nrow(eGOseqs.list2.summary)
#View(eGOseqs.list2.summary)Drawing heatmap
library(scales) # for mute in plot##
## Attaching package: 'scales'## The following object is masked from 'package:purrr':
##
## discard## The following object is masked from 'package:readr':
##
## col_factor# eGOseqs.list2.summary (more than three genes in each KO)
eGOseqs.list2.summary<-eGOseqs.list2.summary[eGOseqs.list2.summary$numDEInCat>3,]
# focused on only very significant GO terms
eGOseqs.list2.summary<-eGOseqs.list2.summary[eGOseqs.list2.summary$over_represented_padjust<1e-4,]
# how to cluster GO Ontology according to this pattern?
## using hclust to sort GO terms
GO.list<-unique(eGOseqs.list2.summary$category)
sample.list<-unique(eGOseqs.list2.summary$sample_FC) # revised
# having x-label also used the x-label for df below
eGOseqs.list2.summary <- eGOseqs.list2.summary %>% mutate(sample=str_remove(sample,".csv")) %>% mutate(sample=str_remove(sample,".DEGs.int")) %>% unite(sample_FC,sample,FC)
# Making matrix for calculate hierarchical clustering of "over-representd_padjust" value using sperad
df<-eGOseqs.list2.summary %>% dplyr::select(category,sample_FC,over_represented_padjust) %>% spread(sample_FC,over_represented_padjust,-1)
df[df<1e-100]<-1e-100 # to avoid "Inf" after log10 transformation
df<- df %>% mutate_at(vars(2:11) ,.funs=function(x) -log10(x))
# df<- df %>% mutate_at(vars(2:11) ,.funs=-log10) # does not work
df[is.na(df)]<-1
df2<-df[,2:11]
rownames(df2)<-df$category
hc<-stats::hclust(dist(df2), "ave") # only numbers
hc$order## [1] 18 51 53 41 5 35 10 52 2 23 25 47 3 30 1 4 31 55 22 39 24 33 14
## [24] 50 48 6 8 34 17 13 15 7 43 9 46 49 32 56 16 38 12 29 45 40 42 44
## [47] 21 37 26 36 27 54 20 28 11 19hc.tib<-tibble(category=hc$labels,order=hc$order)
# change term order using hclust() results
hc.tib2<-hc.tib %>% inner_join(eGOseqs.list2.summary[,c("category","term")],by="category") %>% distinct() %>% arrange(desc(order))
eGOseqs.list2.summary$term<-factor(eGOseqs.list2.summary$term,levels=as_vector(hc.tib2$term))
# To avoid gray color, add 1e-20 to smaller padjust value
eGOseqs.list2.summary$over_represented_padjust[eGOseqs.list2.summary$over_represented_padjust<1e-50]<-1e-50
# format x label
#eGOseqs.list2.summary <- eGOseqs.list2.summary %>% mutate(sample2=str_replace(sample,"trt.density","trt_density")) %>% separate(sample2,into=c("tissue","factor","DEGs","model","ref_density","ref_soil"),sep="\\.") %>% mutate(ref_soil=str_remove(ref_soil,"r")) %>% mutate(ref_density=str_remove(ref_density,"r")) %>% unite(density_FC,ref_density,FC) # simplify elements in "density_FC" column for simpler x label in heatmap
# plot
GOseq.plot<-ggplot(eGOseqs.list2.summary,aes(x=sample_FC,y=term)) + geom_tile(aes(fill=-log10(over_represented_padjust)),colour="black") + scale_fill_gradient2(limit=c(0,50),low=muted("green"), high=muted("magenta")) # OK
GOseq.plot<-GOseq.plot+ theme(axis.text.x=element_text(size=10,angle=90),
axis.text.y=element_text(size=10),
axis.title=element_text(size=10),
axis.ticks = element_blank(),
panel.background = element_rect(fill = "white",colour="black"),
plot.title=element_text(size=20),
axis.line=element_blank()) + labs(x="",y="",title="",fill="-log10\n pvalue")
GOseq.plot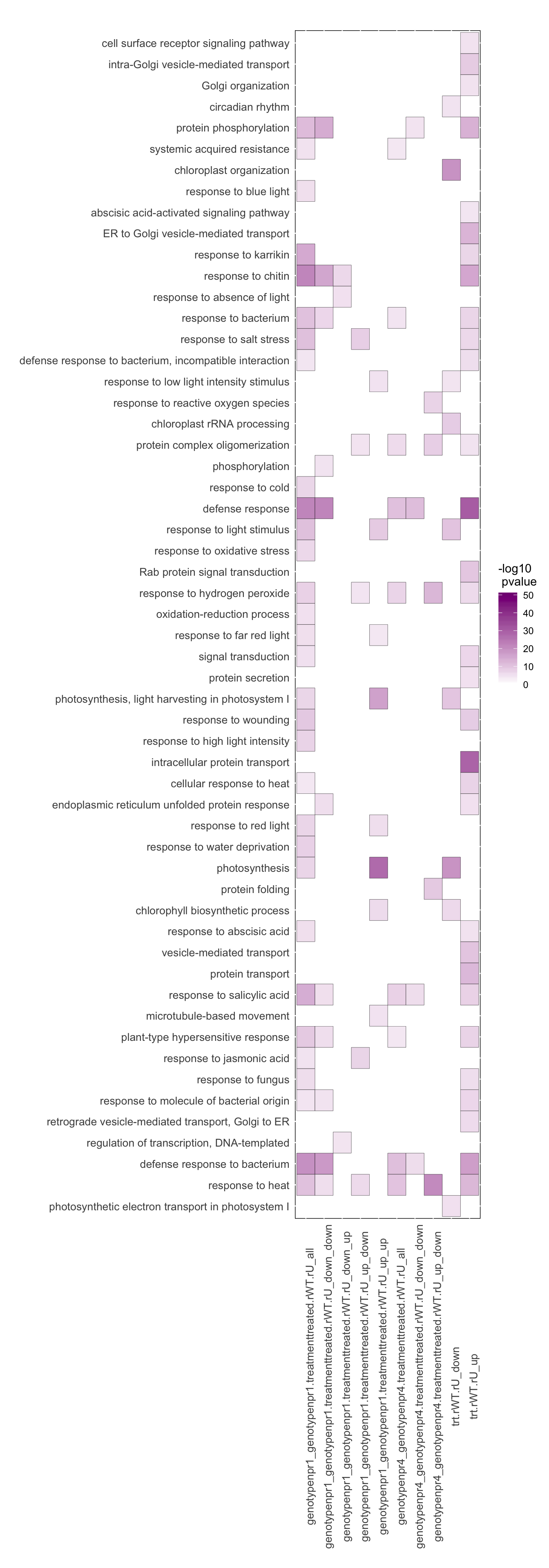
# ggplot_build(GOseq.plot)Session info
sessionInfo()## R version 3.5.1 (2018-07-02)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS High Sierra 10.13.6
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats4 parallel stats graphics grDevices utils datasets
## [8] methods base
##
## other attached packages:
## [1] scales_1.0.0 bindrcpp_0.2.2
## [3] annotate_1.58.0 XML_3.98-1.16
## [5] GO.db_3.6.0 AnnotationDbi_1.42.1
## [7] goseq_1.32.0 geneLenDataBase_1.16.0
## [9] BiasedUrn_1.07 ShortRead_1.38.0
## [11] GenomicAlignments_1.16.0 SummarizedExperiment_1.10.1
## [13] DelayedArray_0.6.6 matrixStats_0.54.0
## [15] Biobase_2.40.0 Rsamtools_1.32.3
## [17] GenomicRanges_1.32.7 GenomeInfoDb_1.16.0
## [19] Biostrings_2.48.0 XVector_0.20.0
## [21] IRanges_2.14.12 S4Vectors_0.18.3
## [23] BiocParallel_1.14.2 BiocGenerics_0.26.0
## [25] forcats_0.3.0 stringr_1.3.1
## [27] dplyr_0.7.7 purrr_0.2.5
## [29] readr_1.1.1 tidyr_0.8.2
## [31] tibble_1.4.2 ggplot2_3.1.0
## [33] tidyverse_1.2.1
##
## loaded via a namespace (and not attached):
## [1] nlme_3.1-137 bitops_1.0-6 lubridate_1.7.4
## [4] bit64_0.9-7 progress_1.2.0 RColorBrewer_1.1-2
## [7] httr_1.3.1 rprojroot_1.3-2 tools_3.5.1
## [10] backports_1.1.2 utf8_1.1.4 R6_2.3.0
## [13] mgcv_1.8-25 DBI_1.0.0 lazyeval_0.2.1
## [16] colorspace_1.3-2 withr_2.1.2 prettyunits_1.0.2
## [19] tidyselect_0.2.5 bit_1.1-14 compiler_3.5.1
## [22] cli_1.0.1 rvest_0.3.2 xml2_1.2.0
## [25] labeling_0.3 rtracklayer_1.40.6 bookdown_0.7
## [28] digest_0.6.18 rmarkdown_1.10 pkgconfig_2.0.2
## [31] htmltools_0.3.6 rlang_0.3.0.1 readxl_1.1.0
## [34] rstudioapi_0.8 RSQLite_2.1.1 bindr_0.1.1
## [37] hwriter_1.3.2 jsonlite_1.5 RCurl_1.95-4.11
## [40] magrittr_1.5 GenomeInfoDbData_1.1.0 Matrix_1.2-15
## [43] fansi_0.4.0 Rcpp_0.12.19 munsell_0.5.0
## [46] stringi_1.2.4 yaml_2.2.0 zlibbioc_1.26.0
## [49] plyr_1.8.4 grid_3.5.1 blob_1.1.1
## [52] crayon_1.3.4 lattice_0.20-35 haven_1.1.2
## [55] GenomicFeatures_1.32.3 hms_0.4.2 knitr_1.20
## [58] pillar_1.3.0 biomaRt_2.36.1 glue_1.3.0
## [61] evaluate_0.12 blogdown_0.9 latticeExtra_0.6-28
## [64] modelr_0.1.2 cellranger_1.1.0 gtable_0.2.0
## [67] assertthat_0.2.0 xfun_0.4 xtable_1.8-3
## [70] broom_0.5.0 memoise_1.1.0