Annotating sequences is critical for making a construct in molecular biology. Benchling.com is a powerful tool for automating the annotation process. This example explains how to label “2x35S promoter” region in sequences of interest.
step1: Before adding 2x35S promoter
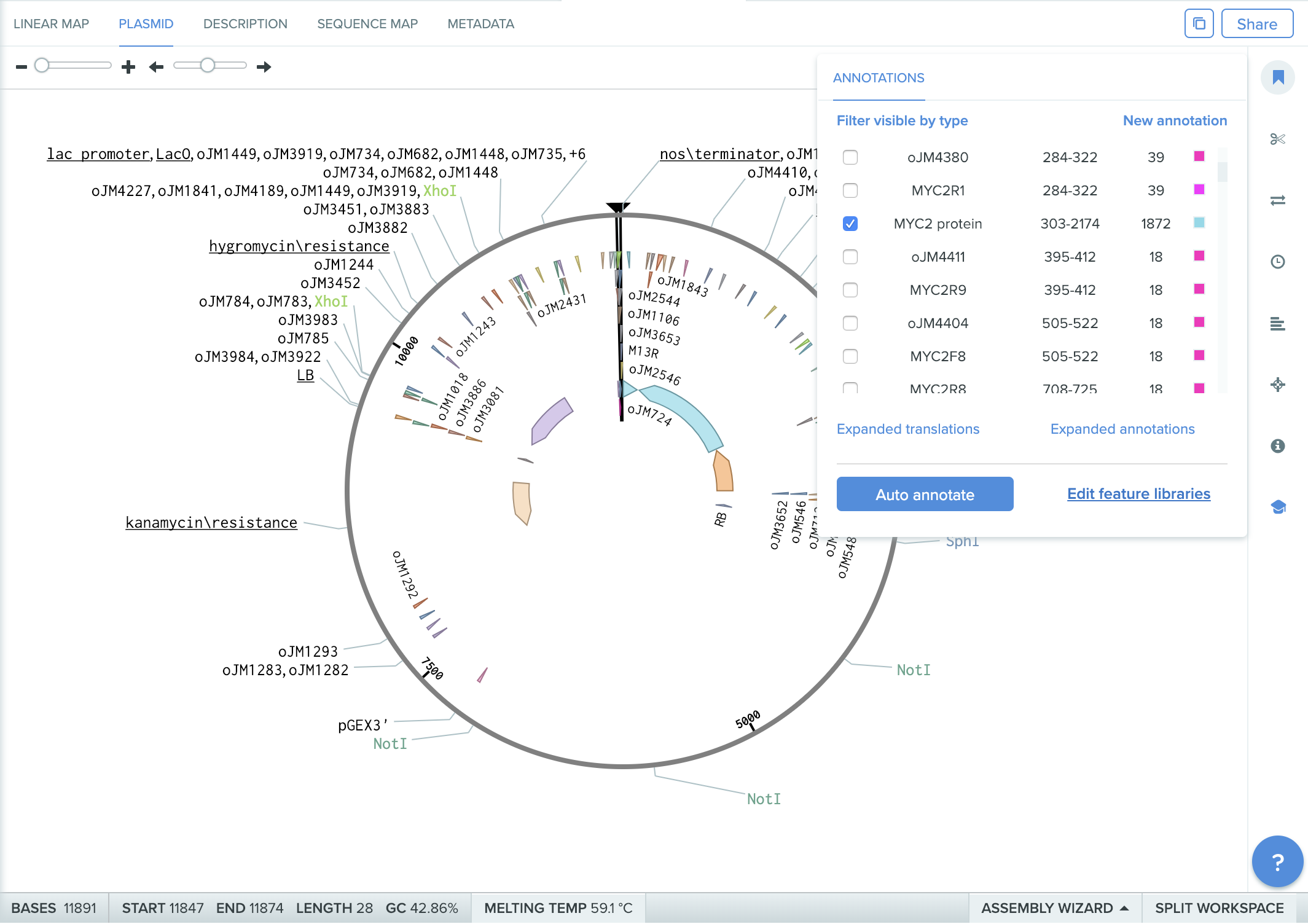
benchling.com plasmid annotation (step1)
step2: Exporting 2x25S promoter feature info (sequences) from known annotated sequence
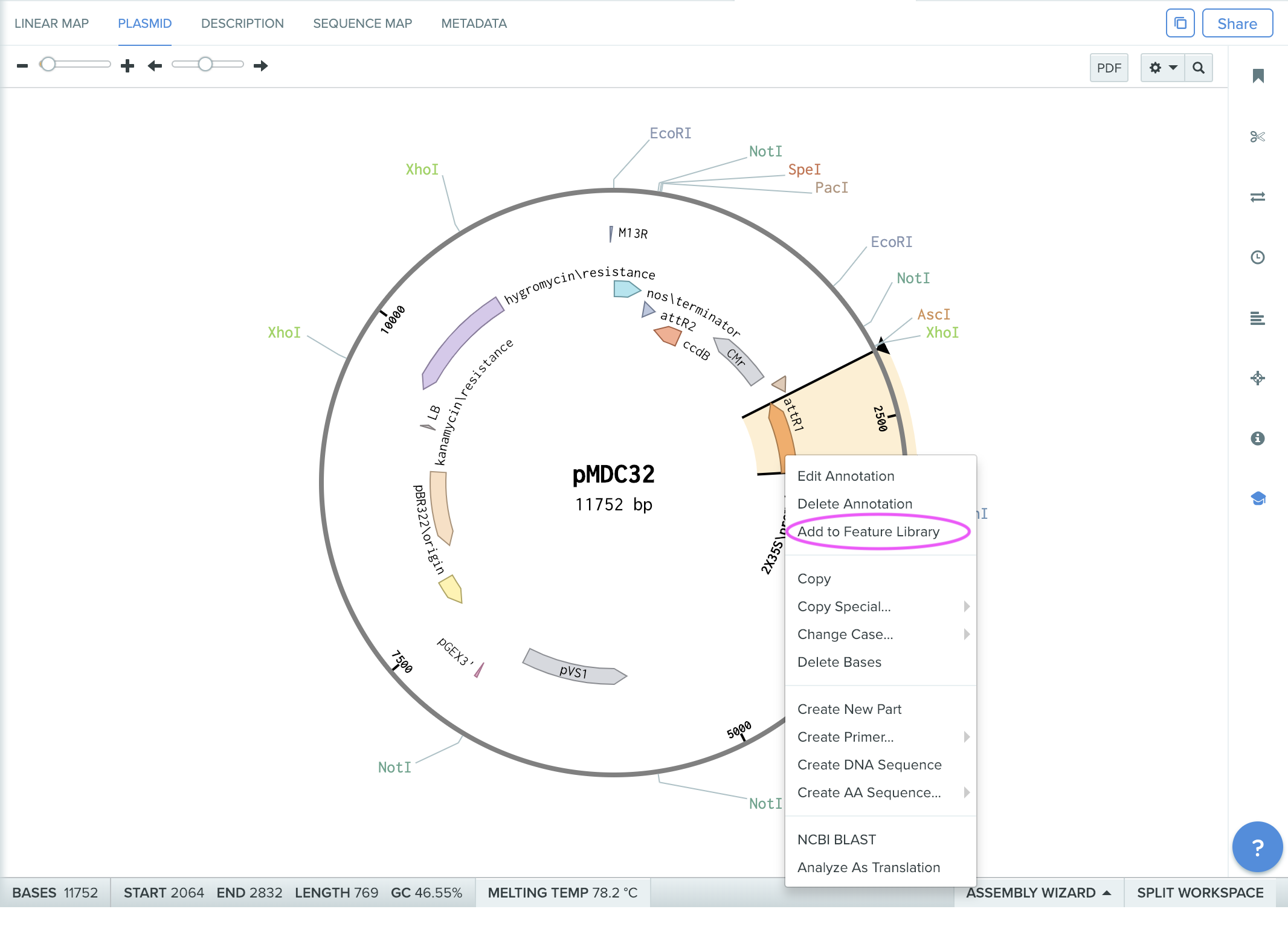 From pMDC32 vector (ref), which has been imported into benchling.com inventory, select “2x25S promoter” feature from “plasmid” view, and then right click to show a window to “Add to Feature Library”.
From pMDC32 vector (ref), which has been imported into benchling.com inventory, select “2x25S promoter” feature from “plasmid” view, and then right click to show a window to “Add to Feature Library”.
step3: Adding “2x25S promoter” to Exisiting Feature Library.
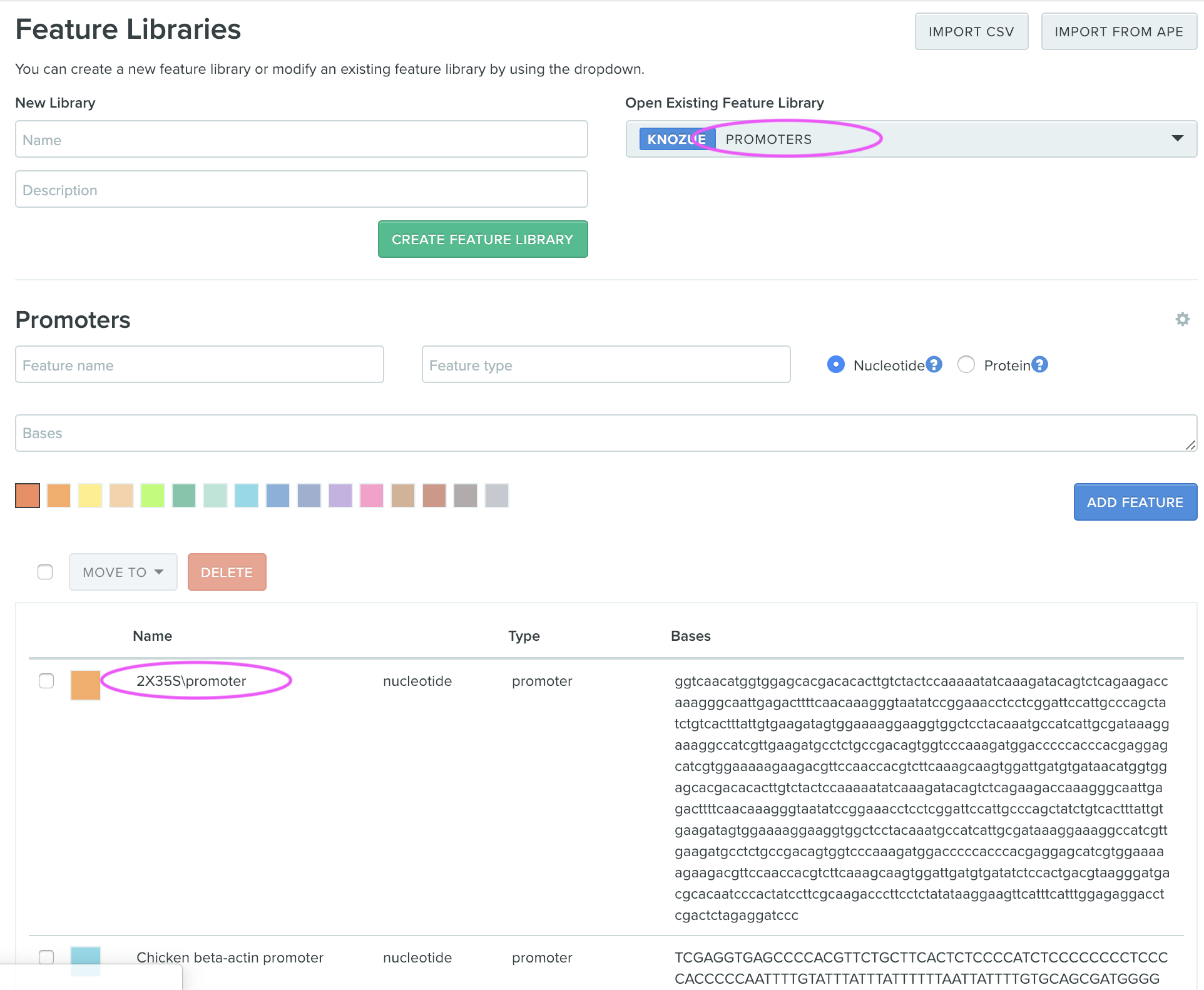 Move to “Feature Libraries” and check if “2x35S promoter” feature has been imported into “PROMOTERS” feature library.
Move to “Feature Libraries” and check if “2x35S promoter” feature has been imported into “PROMOTERS” feature library.
step4: Auto-annotating sequences of interest
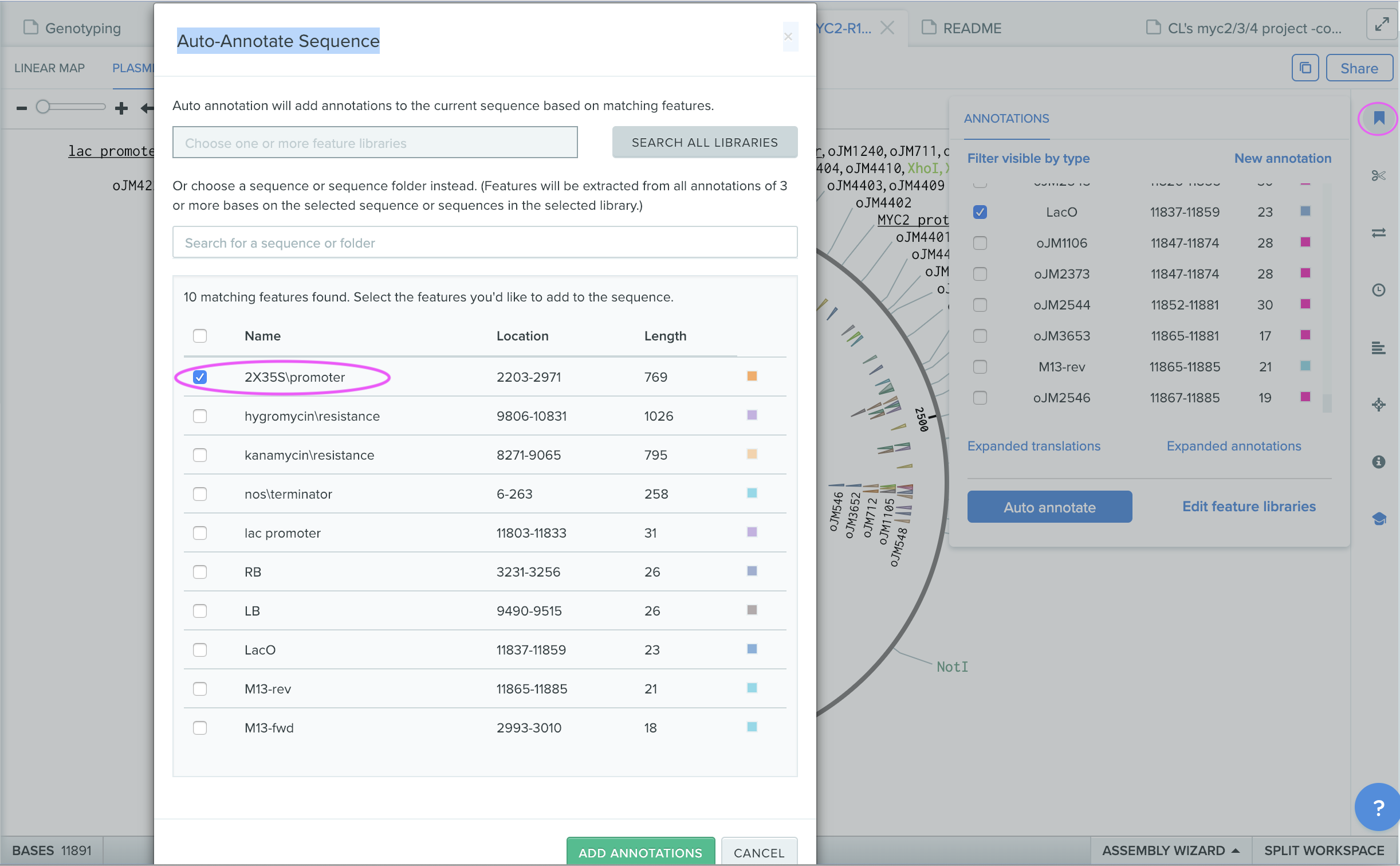 Open a sequence inventory and click an annotation mark at right corner. “Annotations” window will open and then click “Auto annotate”. Results of auto annotated feature list will be open, where “2x25 promoter” has been annotated in the sequences of interest.
Open a sequence inventory and click an annotation mark at right corner. “Annotations” window will open and then click “Auto annotate”. Results of auto annotated feature list will be open, where “2x25 promoter” has been annotated in the sequences of interest.
step5: Visalizing position of the feature in “plasmid” map.
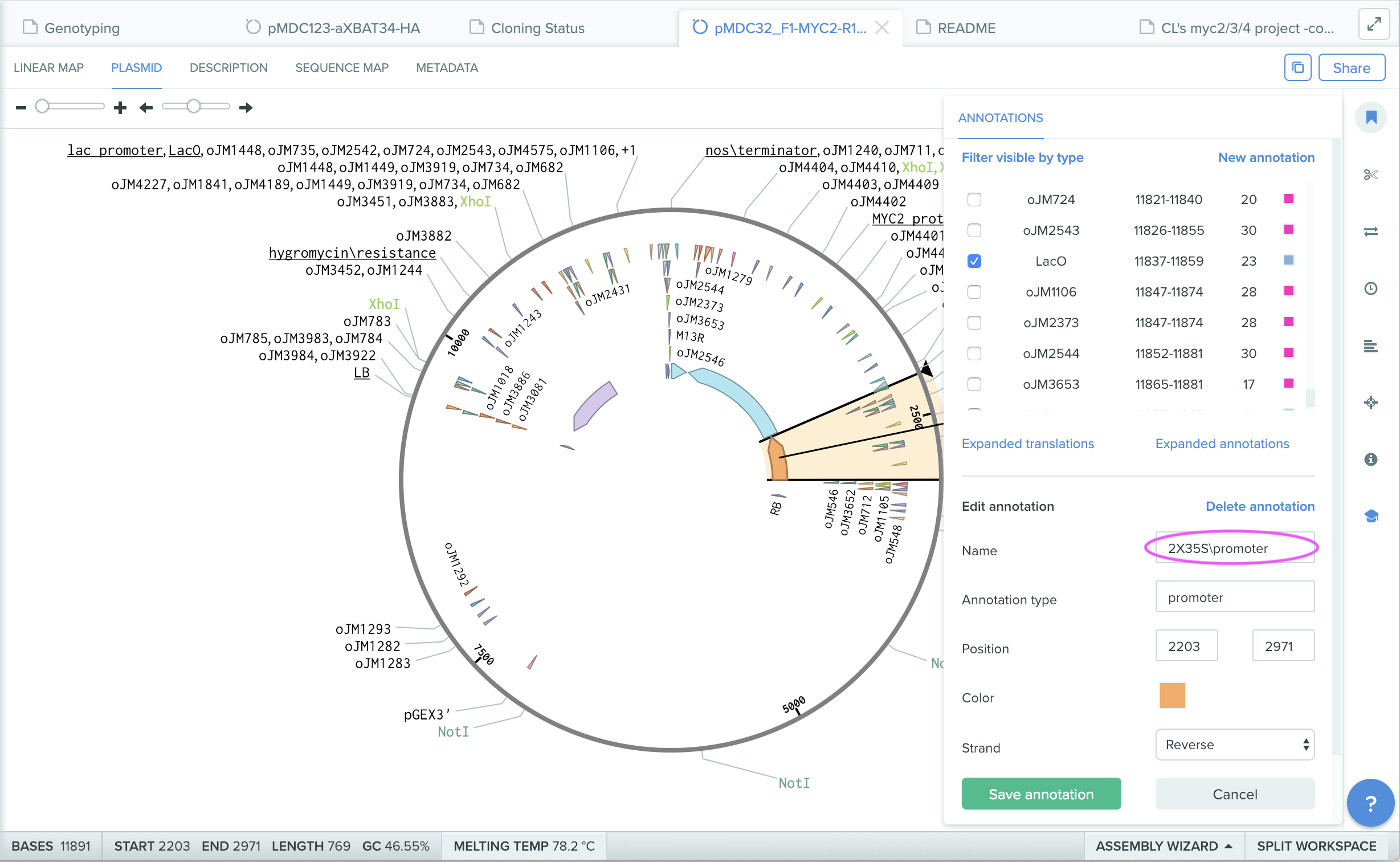 Position of the feature is visualized in “plasmid” map in orange bar. Information of the feature is seen in “Annotations” window. You can repeatedly use the feature library for other sequences.
Position of the feature is visualized in “plasmid” map in orange bar. Information of the feature is seen in “Annotations” window. You can repeatedly use the feature library for other sequences.