How to draw VennDiagram from differentially expressed genes?
The last time I posted how to draw a VennDiagram with a simulated data. This time I would like to explain how to draw a VennDiagram from differentially expressed genes (DEGs).
Overlap between DEGs from Oct 1, 2018 blog post, “Differential expression analysis with public available sequencing data”
library(limma)
library(tidyverse)## ── Attaching packages ───────────────────────────────────────────────────────────────── tidyverse 1.2.1 ──## ✔ ggplot2 3.1.0 ✔ purrr 0.2.5
## ✔ tibble 1.4.2 ✔ dplyr 0.7.7
## ✔ tidyr 0.8.2 ✔ stringr 1.3.1
## ✔ readr 1.1.1 ✔ forcats 0.3.0## ── Conflicts ──────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()# library(ggforce)misregulated geens in npr1-1 in treated samples
load(file.path("2018-12-28-venndiagram-with-differentially-expressed-genes_files","dge.nolow.Ding2018.Rdata"))
dge.nolow## Loading required package: edgeR## An object of class "DGEList"
## $counts
## SRR6739807 SRR6739808 SRR6739809 SRR6739810 SRR6739811 SRR6739812
## 1 3598.000 3442.000 3653 3531 4027 3775
## 4 427.000 412.000 316 348 319 357
## 5 132.805 129.168 154 141 155 131
## 6 52.000 46.000 74 110 122 91
## 7 685.000 764.000 231 220 264 247
## SRR6739813 SRR6739814 SRR6739815 SRR6739816 SRR6739817 SRR6739818
## 1 4195 4317.9900 4015.000 3874.000 3663.000 3642
## 4 286 306.0000 389.000 362.000 290.000 285
## 5 126 99.8598 156.895 130.783 101.738 120
## 6 118 135.0000 60.000 52.000 85.000 79
## 7 118 131.0000 416.000 383.000 104.000 164
## 16794 more rows ...
##
## $samples
## group lib.size norm.factors Run
## SRR6739807 WT_treated 21747803 0.9813040 SRR6739807
## SRR6739808 WT_treated 21005249 0.9969243 SRR6739808
## SRR6739809 WT_untreated 21052743 0.9901012 SRR6739809
## SRR6739810 WT_untreated 22518666 0.9893923 SRR6739810
## SRR6739811 npr1-1_treated 21437014 1.0185940 SRR6739811
## sample group.1 genotype treatment rep
## SRR6739807 WT_treated_1 WT_treated WT treated 1
## SRR6739808 WT_treated_2 WT_treated WT treated 2
## SRR6739809 WT_untreated_1 WT_untreated WT untreated 1
## SRR6739810 WT_untreated_2 WT_untreated WT untreated 2
## SRR6739811 npr1-1_treated_1 npr1-1_treated npr1-1 treated 1
## LibraryLayout
## SRR6739807 SINGLE
## SRR6739808 SINGLE
## SRR6739809 SINGLE
## SRR6739810 SINGLE
## SRR6739811 SINGLE
## 7 more rows ...
##
## $genes
## genes
## 1 AT1G50920.1
## 4 AT1G15970.1
## 5 AT1G73440.1
## 6 AT1G75120.1
## 7 AT1G17600.1
## 16794 more rows ...# set references in genotype and treatment
dge.nolow$samples$genotype<-factor(dge.nolow$samples$genotype,levels=c("WT","npr4-4D","npr1-1"))
dge.nolow$samples$treatment<-factor(dge.nolow$samples$treatment,levels=c("treated","untreated"))
# making model matrix
design.int <- with(dge.nolow$samples, model.matrix(~ genotype*treatment + rep))
# estimateDisp
dge.nolow.int <- estimateDisp(dge.nolow,design = design.int)
## fit linear model
fit.int <- glmFit(dge.nolow.int,design.int)
# get DEGs, genotype effects in npr1-1 (no matter treated or untreated) (coef=c("genotypenpr1-1","genotypenpr1-1:treatmenttreated"))
genotypenpr1_genotypenpr1.treatmentuntreated.lrt.rWT.rT <- glmLRT(fit.int,coef = c("genotypenpr1-1","genotypenpr1-1:treatmentuntreated"))
genotypenpr1_genotypenpr1.treatmentuntreated.DEGs.int.rWT.rT <- topTags(genotypenpr1_genotypenpr1.treatmentuntreated.lrt.rWT.rT,n = Inf,p.value = 0.05)$table;nrow(genotypenpr1_genotypenpr1.treatmentuntreated.DEGs.int.rWT.rT) # ## [1] 6460Download updated gene description
Most updated annotation can be downloaded from TAIR website (for subscribers). I used “TAIR Data 20180630”.
At.gene.name <-read_tsv("https://www.arabidopsis.org/download_files/Subscriber_Data_Releases/TAIR_Data_20180630/gene_aliases_20180702.txt.gz") # Does work from home when I use Pulse Secure.## Parsed with column specification:
## cols(
## name = col_character(),
## symbol = col_character(),
## full_name = col_character()
## )# test %>% dplyr::select(name,symbol) %>% group_by(name) %>% spread(key=name,value=symbol)
# test %>% dplyr::select(name,symbol) %>% spread(key=symbol,value=name)
# test %>% dplyr::select(name,symbol) %>% group_by(name) %>% spread(key=symbol,value=name)
# test %>% dplyr::select(name,symbol) %>% group_by(name) %>% summarise(n())
# How to concatenate multiple symbols in the same AGI
At.gene.name <- At.gene.name %>% group_by(name) %>% summarise(symbol2=paste(symbol,collapse=";"),full_name=paste(full_name,collapse=";"))
At.gene.name %>% dplyr::slice(100:110) # OK## # A tibble: 11 x 3
## name symbol2 full_name
## <chr> <chr> <chr>
## 1 AT1G018… AtGEN1;GEN1 NA;ortholog of HsGEN1
## 2 AT1G019… ATSBT1.1;SBTI1.1 NA;NA
## 3 AT1G019… AtGET3a;GET3a Guided Entry of Tail-anchored protein 3a;Gui…
## 4 AT1G019… ARK2;AtKINUb armadillo repeat kinesin 2;Arabidopsis thali…
## 5 AT1G019… BIG3;EDA10 BIG3;embryo sac development arrest 10
## 6 AT1G019… AtBBE1;OGOX4 NA;oligogalacturonide oxidase 4
## 7 AT1G020… GAE2 UDP-D-glucuronate 4-epimerase 2
## 8 AT1G020… SEC1A secretory 1A
## 9 AT1G020… LAP6;PKSA LESS ADHESIVE POLLEN 6;polyketide synthase A
## 10 AT1G020… SPL8 squamosa promoter binding protein-like 8
## 11 AT1G020… ATCSN7;COP15;CS… ARABIDOPSIS THALIANA COP9 SIGNALOSOME SUBUNI…Add gene descripion to DEG files
# npr1-1 (either treated or untreated)
genotypenpr1_genotypenpr1.treatmentuntreated.DEGs.int.rWT.rT.desc <- genotypenpr1_genotypenpr1.treatmentuntreated.DEGs.int.rWT.rT %>% mutate(AGI=str_remove(genes,pattern="\\.[[:digit:]]+$")) %>% left_join(At.gene.name,by=c("AGI"="name"))
write_csv(genotypenpr1_genotypenpr1.treatmentuntreated.DEGs.int.rWT.rT.desc,"2018-12-28-venndiagram-with-differentially-expressed-genes_files/output/genotypenpr1_genotypenpr1.treatmentuntreated.DEGs.int.rWT.rT.csv")read DEG files
# read csv files
getwd()## [1] "/Volumes/data_personal/Kazu_blog/content/post"## read csv file names
DEG.objs<-list.files(path=file.path("2018-12-28-venndiagram-with-differentially-expressed-genes_files","output"))
DEG.objs## [1] "genotypenpr1_genotypenpr1.treatmenttreated.DEGs.int.rWT.rU.csv"
## [2] "genotypenpr1_genotypenpr1.treatmentuntreated.DEGs.int.rWT.rT.csv"
## [3] "genotypenpr4_genotypenpr4.treatmenttreated.DEGs.int.rWT.rU.csv"
## [4] "trt.DEGs.int.rWT.rU.csv"## read csv files
DEG.count.list<-lapply(DEG.objs, function(x) read_csv(paste(file.path("2018-12-28-venndiagram-with-differentially-expressed-genes_files","output"),"/",x,sep="")))## Parsed with column specification:
## cols(
## genes = col_character(),
## logFC.genotypenpr1.1 = col_double(),
## logFC.genotypenpr1.1.treatmenttreated = col_double(),
## logCPM = col_double(),
## LR = col_double(),
## PValue = col_double(),
## FDR = col_double(),
## AGI = col_character(),
## symbol2 = col_character(),
## full_name = col_character()
## )## Parsed with column specification:
## cols(
## genes = col_character(),
## logFC.genotypenpr1.1 = col_double(),
## logFC.genotypenpr1.1.treatmentuntreated = col_double(),
## logCPM = col_double(),
## LR = col_double(),
## PValue = col_double(),
## FDR = col_double(),
## AGI = col_character(),
## symbol2 = col_character(),
## full_name = col_character()
## )## Parsed with column specification:
## cols(
## genes = col_character(),
## logFC.genotypenpr4.4D = col_double(),
## logFC.genotypenpr4.4D.treatmenttreated = col_double(),
## logCPM = col_double(),
## LR = col_double(),
## PValue = col_double(),
## FDR = col_double(),
## AGI = col_character(),
## symbol2 = col_character(),
## full_name = col_character()
## )## Parsed with column specification:
## cols(
## genes = col_character(),
## logFC = col_double(),
## logCPM = col_double(),
## LR = col_double(),
## PValue = col_double(),
## FDR = col_double(),
## AGI = col_character(),
## symbol2 = col_character(),
## full_name = col_character()
## )names(DEG.count.list)<-gsub(".csv","",DEG.objs)Interpretation of edgeR glm double coefficient results (topTag)
names(DEG.count.list)## [1] "genotypenpr1_genotypenpr1.treatmenttreated.DEGs.int.rWT.rU"
## [2] "genotypenpr1_genotypenpr1.treatmentuntreated.DEGs.int.rWT.rT"
## [3] "genotypenpr4_genotypenpr4.treatmenttreated.DEGs.int.rWT.rU"
## [4] "trt.DEGs.int.rWT.rU"# reference in treatment was "untreated"
DEG.count.list[["genotypenpr1_genotypenpr1.treatmenttreated.DEGs.int.rWT.rU"]]## # A tibble: 6,460 x 10
## genes logFC.genotypen… logFC.genotypen… logCPM LR PValue FDR
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 AT2G… -4.19 -0.733 6.69 1327. 6.75e-289 1.13e-284
## 2 AT5G… 3.26 0.517 4.76 1072. 1.33e-233 1.11e-229
## 3 AT1G… -2.01 -0.488 7.19 659. 6.38e-144 3.57e-140
## 4 AT1G… -2.25 -0.525 7.51 624. 2.54e-136 1.07e-132
## 5 AT3G… -3.15 0.874 5.33 622. 1.00e-135 3.36e-132
## 6 AT5G… -0.206 -6.73 3.90 595. 7.88e-130 2.21e-126
## 7 AT1G… -3.64 -1.53 3.37 581. 7.28e-127 1.75e-123
## 8 AT5G… 1.83 1.82 9.40 571. 1.04e-124 2.18e-121
## 9 AT2G… 2.23 1.94 9.22 559. 4.26e-122 7.94e-119
## 10 AT5G… -2.01 -3.49 4.19 538. 1.19e-117 2.00e-114
## # ... with 6,450 more rows, and 3 more variables: AGI <chr>,
## # symbol2 <chr>, full_name <chr># reference in treatment was "treated"
## genotypenpr1_genotypenpr1.treatmentuntreated.DEGs.int.rWT.rT
DEG.count.list[["genotypenpr1_genotypenpr1.treatmentuntreated.DEGs.int.rWT.rT"]]## # A tibble: 6,460 x 10
## genes logFC.genotypen… logFC.genotypen… logCPM LR PValue FDR
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 AT2G… -4.92 0.733 6.69 1327. 6.75e-289 1.13e-284
## 2 AT5G… 3.78 -0.517 4.76 1072. 1.33e-233 1.11e-229
## 3 AT1G… -2.50 0.488 7.19 659. 6.38e-144 3.57e-140
## 4 AT1G… -2.78 0.525 7.51 624. 2.54e-136 1.07e-132
## 5 AT3G… -2.28 -0.874 5.33 622. 1.00e-135 3.36e-132
## 6 AT5G… -6.94 6.73 3.90 595. 7.88e-130 2.21e-126
## 7 AT1G… -5.17 1.53 3.37 581. 7.28e-127 1.75e-123
## 8 AT5G… 3.65 -1.82 9.40 571. 1.04e-124 2.18e-121
## 9 AT2G… 4.17 -1.94 9.22 559. 4.26e-122 7.94e-119
## 10 AT5G… -5.50 3.49 4.19 538. 1.19e-117 2.00e-114
## # ... with 6,450 more rows, and 3 more variables: AGI <chr>,
## # symbol2 <chr>, full_name <chr># Total numbe of DEGS are the same off course.
# Absolute value of "logFC.genotypenpr1.1.treatmenttreated" in genotypenpr1_genotypenpr1.treatmenttreated.DEGs.int.rWT.rU.csv is the same with one in "logFC.genotypenpr1.1.treatmentuntreated" in genotypenpr1_genotypenpr1.treatmentuntreated.DEGs.int.rWT.rT.csv and only signs (positive or negative) are different.
# logFC.genotypenpr1.1 column
## In "genotypenpr1_genotypenpr1.treatmentuntreated.DEGs.int.rWT.rT.csv", logFC value are calculated for "treated" (=reference is "treated"; rT) samples.
## In "genotypenpr1_genotypenpr1.treatmenttreated.DEGs.int.rWT.rU.csv", logFC value are calculated for "treated" (=reference is "treated"; rT) samples.making summary table
# combine all DEG gene names
all.DEGs<-unique(c(DEG.count.list[["genotypenpr1_genotypenpr1.treatmenttreated.DEGs.int.rWT.rU"]]$genes,DEG.count.list[["trt.DEGs.int.rWT.rU"]]$genes))
#
WT.SA.up<-DEG.count.list[["trt.DEGs.int.rWT.rU"]] %>% filter(logFC>0) %>% mutate(WT.SA.up=1) %>% select(genes,WT.SA.up)
WT.SA.down<-DEG.count.list[["trt.DEGs.int.rWT.rU"]] %>% filter(logFC<0) %>% mutate(WT.SA.down=1) %>% select(genes,WT.SA.down)
# npr1.1 regulated genes
npr1.gt<-DEG.count.list[["genotypenpr1_genotypenpr1.treatmenttreated.DEGs.int.rWT.rU"]] %>% mutate(npr1.gt=1) %>% select(genes,npr1.gt)
# summarize
summary.DEGS<-tibble(genes=all.DEGs) %>% full_join(WT.SA.up,by="genes") %>% full_join(WT.SA.down,by="genes") %>% full_join(npr1.gt,by="genes")
dim(summary.DEGS)[1]==length(all.DEGs)## [1] TRUEsummary.DEGS[is.na(summary.DEGS)]<-0
# convert into count table
vdc<-limma::vennCounts(summary.DEGS[,-1])
# plot (classical way)
limma::vennDiagram(vdc) # traditional version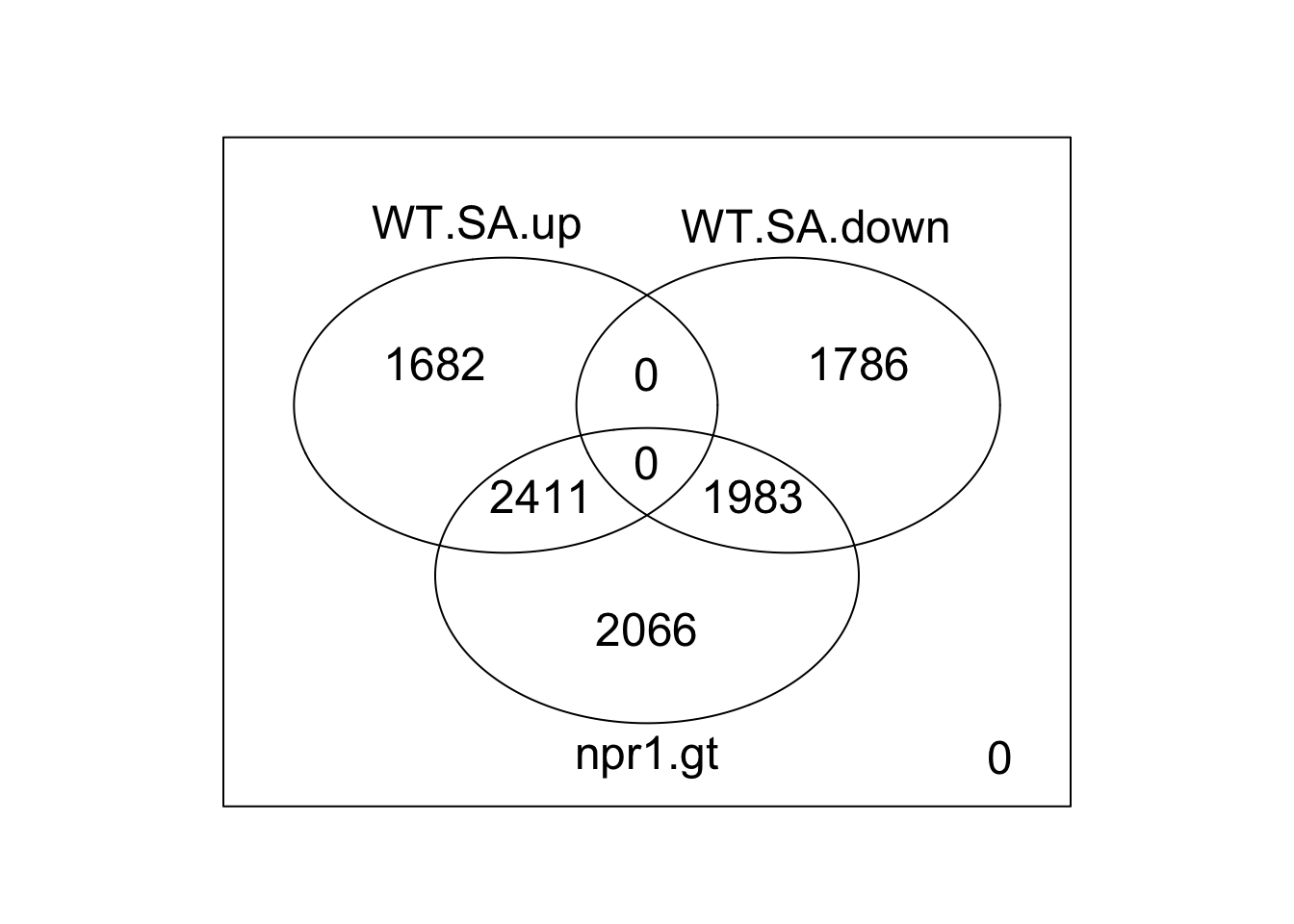
# plot (ggplot2 version) ### under construction ####Session info
sessionInfo()## R version 3.5.1 (2018-07-02)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS 10.14.1
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] bindrcpp_0.2.2 edgeR_3.22.5 forcats_0.3.0 stringr_1.3.1
## [5] dplyr_0.7.7 purrr_0.2.5 readr_1.1.1 tidyr_0.8.2
## [9] tibble_1.4.2 ggplot2_3.1.0 tidyverse_1.2.1 limma_3.36.5
##
## loaded via a namespace (and not attached):
## [1] locfit_1.5-9.1 tidyselect_0.2.5 xfun_0.4 splines_3.5.1
## [5] haven_1.1.2 lattice_0.20-35 colorspace_1.3-2 htmltools_0.3.6
## [9] yaml_2.2.0 utf8_1.1.4 rlang_0.3.0.1 pillar_1.3.0
## [13] glue_1.3.0 withr_2.1.2 modelr_0.1.2 readxl_1.1.0
## [17] bindr_0.1.1 plyr_1.8.4 munsell_0.5.0 blogdown_0.9
## [21] gtable_0.2.0 cellranger_1.1.0 rvest_0.3.2 evaluate_0.12
## [25] knitr_1.21 curl_3.2 fansi_0.4.0 broom_0.5.0
## [29] Rcpp_1.0.0 scales_1.0.0 backports_1.1.2 jsonlite_1.6
## [33] hms_0.4.2 digest_0.6.18 stringi_1.2.4 bookdown_0.7
## [37] grid_3.5.1 cli_1.0.1 tools_3.5.1 magrittr_1.5
## [41] lazyeval_0.2.1 crayon_1.3.4 pkgconfig_2.0.2 xml2_1.2.0
## [45] lubridate_1.7.4 assertthat_0.2.0 rmarkdown_1.11 httr_1.3.1
## [49] rstudioapi_0.8 R6_2.3.0 nlme_3.1-137 compiler_3.5.1