RNAseq is a powerful tool for transcriptome analysis. It is also useful for genotyping (for example, 1). Here I demonstrated an example of confirmation of genotype used for RNAseq. There are some unavoidable errors due to unexpected mistakes in many steps (eg. contamination of seeds, contamination of samples, pipetting errors during library preparation, rotation of plates, etc.). Therefore confirmation of genotype using RNAseq data is useful for quality control of samples used 2. Please find bam files used in my previous blog “Differential expression analysis with public available sequencing data” on Oct 1, 2018. A tool for handling bam files is Samtools.
chop NPR1 cDNA from each bam
setwd("kallisto_sam_out2")
sorted.bam.files<-list.files(pattern="sorted.bam$",recursive = TRUE)
SRRs<-list.files(pattern="(SRR)([[:digit:]]+)$")
for(y in SRRs) {
system(paste("samtools view -b ",y,"/*.sorted.bam AT1G64280.1 -o ",y,"_AT1G64280.bam",sep=""))
} # "AT1G64280" is a gene ID for NPR1 and AT1G64280.1 is a representative splicing variant.
# sample info from NCBI
sample.info<-read_csv("http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?save=efetch&rettype=runinfo&db=sra&term=PRJNA434313") # Directly from ncbi site. This works!
# convert sample info into human readable version
sample.info2<-tibble(SampleName=c("GSM3014771","GSM3014772","GSM3014773","GSM3014774","GSM3014775","GSM3014776","GSM3014777","GSM3014778","GSM3014779","GSM3014780","GSM3014781","GSM3014782","GSM3014783","GSM3014784","GSM3014785","GSM3014786"),sample=c("WT_treated_1","WT_treated_2","WT_untreated_1","WT_untreated_2","npr1-1_treated_1","npr1-1_treated_2","npr1-1_untreated_1","npr1-1_untreated_2","npr4-4D_treated_1","npr4-4D_treated_2","npr4-4D_untreated_1","npr4-4D_untreated_2","npr1-1_npr4-4D_treated_1","npr1-1_npr4-4D_treated_2","npr1-1_npr4-4D_untreated_1","npr1-1_npr4-4D_untreated_2")) # # from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110702
# combine
sample.info.final <- left_join(sample.info,sample.info2,by="SampleName") %>% dplyr::select(Run,sample,LibraryLayout) %>% separate(sample,into=c("genotype1","genotype2","treatment","rep"),sep="_",fill="left",remove=FALSE) %>% unite(genotype,genotype1,genotype2,sep="_") %>% mutate(genotype=str_replace(genotype,"NA_","")) %>% unite(group,genotype,treatment,sep="_",remove=FALSE)index each bam
SRRs_npr1.bam<-list.files(pattern="AT1G64280.bam")
sapply(paste("samtools index -b ",SRRs_npr1.bam,sep=""),function(x) system(x))Visualize npr1-1 mutation by IGV
To visualize sequence reads (also called “reads”) Integrative Genomics Viewer (IGV, v2.4.10) was used. Mutation of C to T that lead a missense mutation (His (CAT) to Tyr (TAT)) in the third ankyrin repeat 3 was confirmed.
Known npr1-1 mutation (from my note on benchling.com)
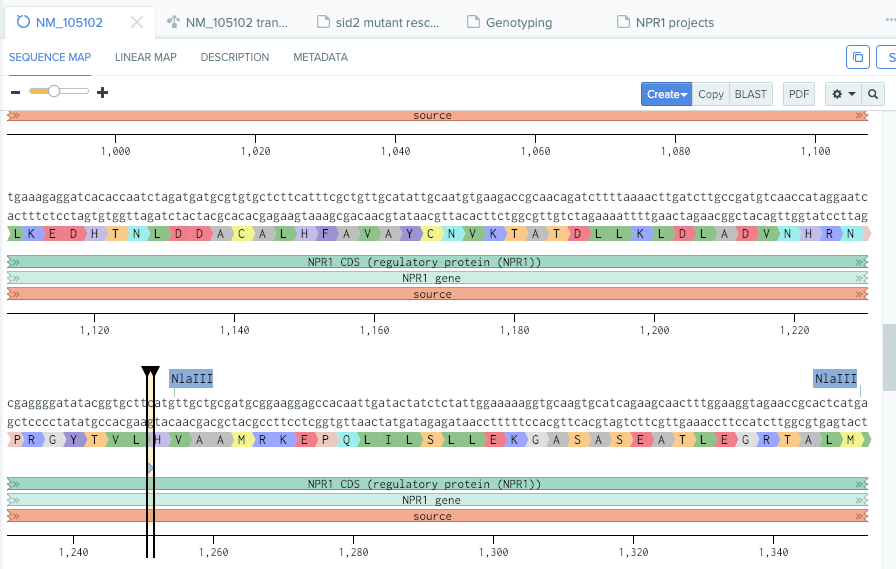
benchling.com npr1-1 mutation
IGV view of NPR1 cDNA
Upper track is wild type and lower track is npr1-1.
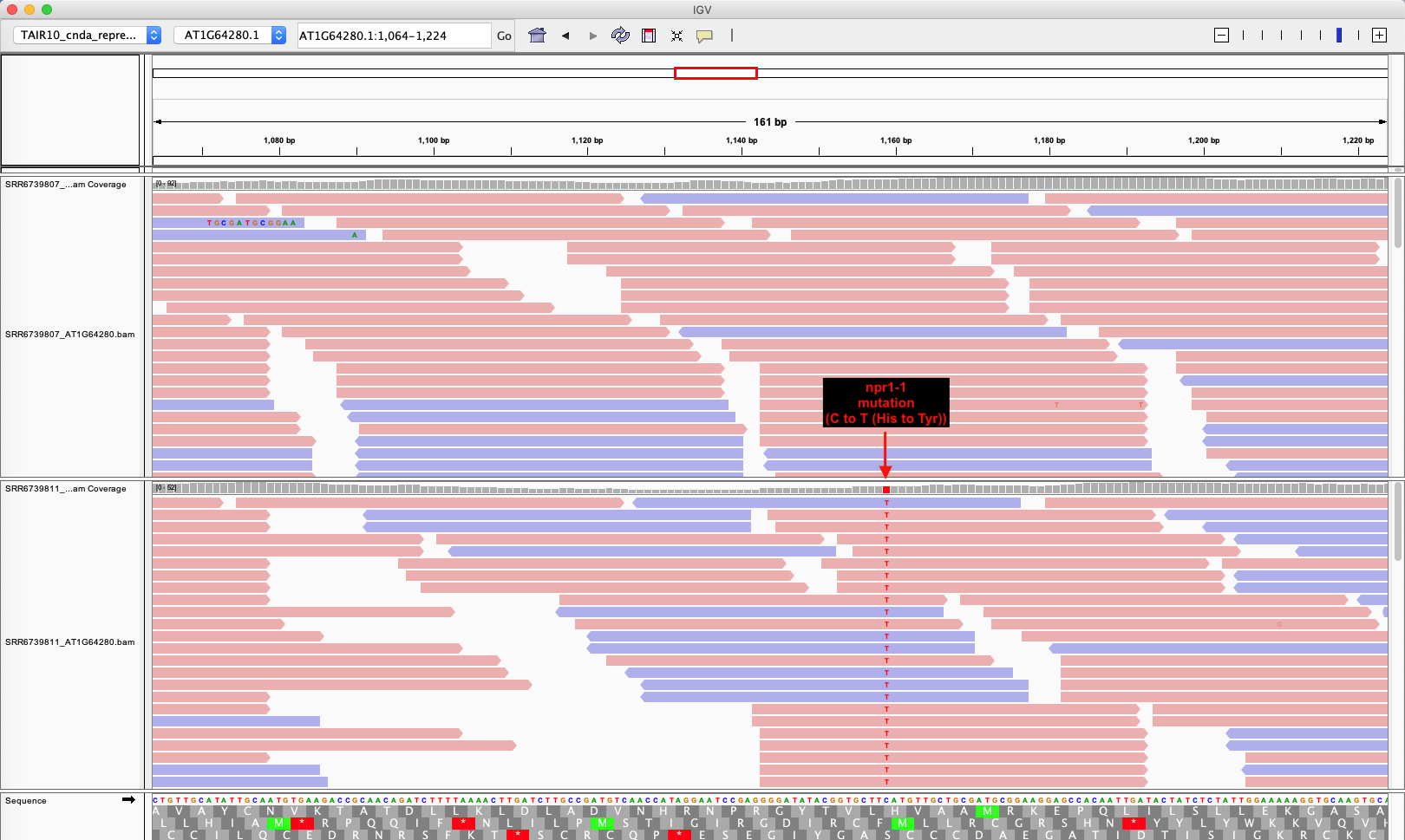
IGV Ding 2018 NPR1
Session info
sessionInfo()## R version 3.5.1 (2018-07-02)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS 10.14.3
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## loaded via a namespace (and not attached):
## [1] compiler_3.5.1 magrittr_1.5 bookdown_0.9 tools_3.5.1
## [5] htmltools_0.3.6 yaml_2.2.0 Rcpp_1.0.0 stringi_1.2.4
## [9] rmarkdown_1.11 blogdown_0.10 knitr_1.21 stringr_1.3.1
## [13] digest_0.6.18 xfun_0.4 evaluate_0.12R books
R and bioinformatics
Koenig D, Jiménez-Gómez JM, Kimura S, Fulop D, Chitwood DH, Headland LR, Kumar R, Covington MF, Devisetty UK, Tat AV, et al (2013) Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. PNAS 110: E2655–E2662↩
Nozue K, Devisetty UK, Lekkala S, Mueller-Moulé P, Bak A, Casteel CL, Maloof JN (2018) Network Analysis Reveals a Role for Salicylic Acid Pathway Components in Shade Avoidance. Plant Physiology 178: 1720–1732↩
Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63↩