library(XML)
library(tidyverse)
library(lubridate)
library(scales)
library(ggthemes)
Reading downloaded health data from an Apple watch.
path="/Volumes/data_personal/Kazu_blog/resources"
#zip <- paste(path, 'export_Mana_101320.zip', sep = '/')
zip <- paste(path, 'export_Mana_122620.zip', sep = '/')
unzip(zip, exdir = path)
Sys.sleep(3) # pause for 3 seconds to let your computer unzip it.
Parse the xml file.
xml <- xmlParse(paste0(path, '/apple_health_export/export.xml'))
summary(xml)
## $nameCounts
##
## Record MetadataEntry
## 625705 57389
## InstantaneousBeatsPerMinute WorkoutEvent
## 6004 489
## ActivitySummary HeartRateVariabilityMetadataList
## 354 182
## Workout FileReference
## 113 47
## WorkoutRoute ExportDate
## 47 1
## HealthData Me
## 1 1
##
## $numNodes
## [1] 690333
Convert xml data into dataframes.
df_record <- XML:::xmlAttrsToDataFrame(xml["//Record"])
df_activity <- XML:::xmlAttrsToDataFrame(xml["//ActivitySummary"])
df_workout <- XML:::xmlAttrsToDataFrame(xml["//Workout"])
# df_clinical <- XML:::xmlAttrsToDataFrame(xml["//ClinicalRecord"])
# df_location <- XML:::xmlAttrsToDataFrame(xml["//Location"]) %>%
# mutate(latitude = as.numeric(as.character(latitude)),
# longitude = as.numeric(as.character(longitude)))
What type of health data we have?
df %>% select(type) %>% distinct()
## type
## 1 Height
## 2 BodyMass
## 3 HeartRate
## 4 StepCount
## 5 DistanceWalkingRunning
## 6 BasalEnergyBurned
## 7 ActiveEnergyBurned
## 8 FlightsClimbed
## 9 AppleExerciseTime
## 10 RestingHeartRate
## 11 WalkingHeartRateAverage
## 12 HeadphoneAudioExposure
## 13 AppleStandTime
## 14 HKCategoryTypeIdentifierAppleStandHour
## 15 HKCategoryTypeIdentifierMenstrualFlow
## 16 HKCategoryTypeIdentifierMindfulSession
## 17 HKCategoryTypeIdentifierBloating
## 18 HKCategoryTypeIdentifierDiarrhea
## 19 HeartRateVariabilitySDNN
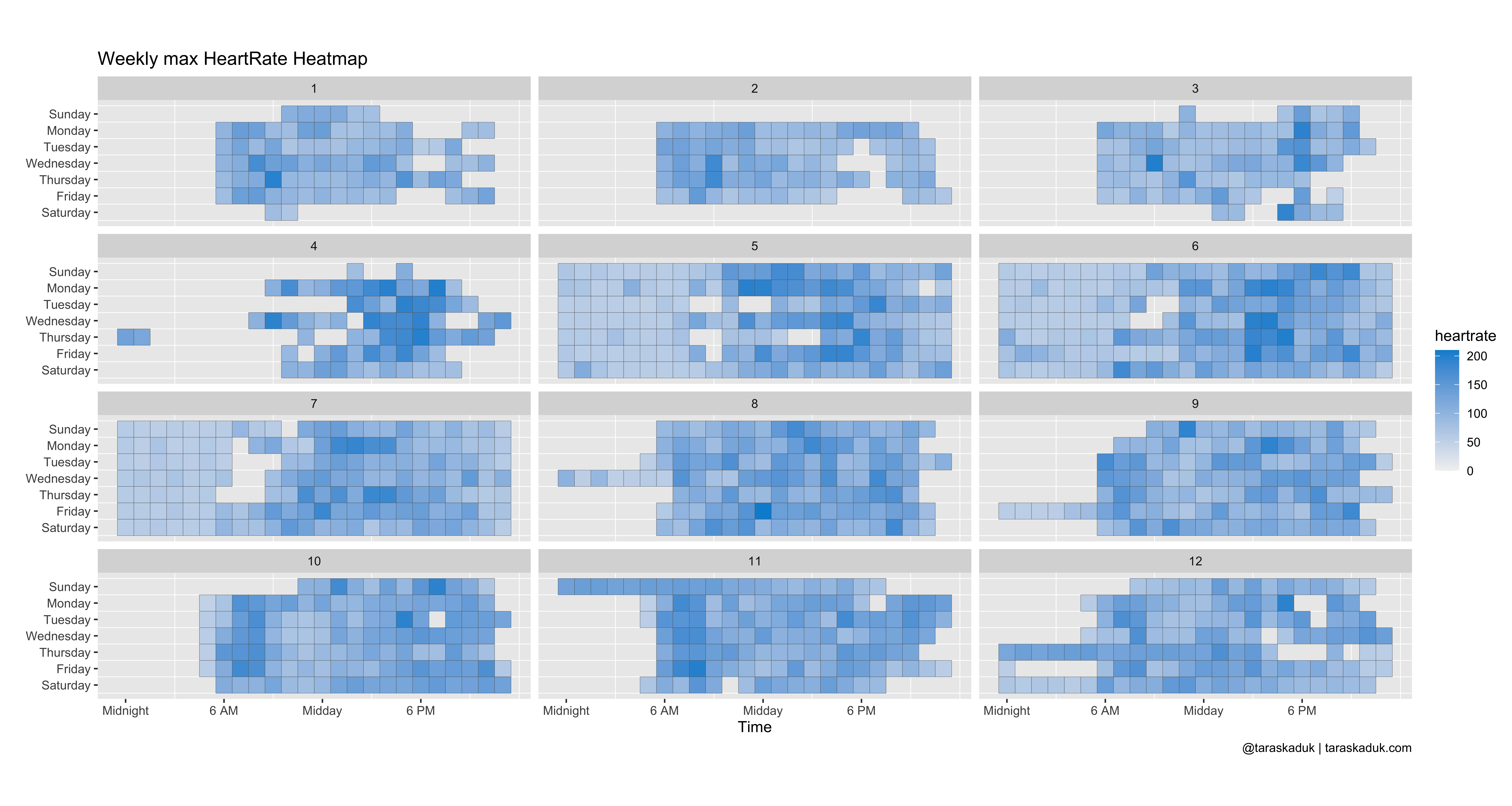
Plot heartRate heatmap (weekly: max)
# revised
df %>%
dplyr::filter(type == 'HeartRate') %>%
dplyr::filter(year==2020) %>%
group_by(month,wday,hour) %>%
summarize(heartrate=max(value)) %>%
arrange(desc(heartrate)) %>%
ggplot(aes(x=hour, y=wday, fill=heartrate)) +
geom_tile(col = 'grey40') +
scale_fill_continuous(limit=c(0,210),labels = scales::comma, low = 'grey95', high = '#008FD5') +
theme(panel.grid.major = element_blank()) +
scale_x_continuous(
breaks = c(0, 6, 12, 18),
label = c("Midnight", "6 AM", "Midday", "6 PM")
) +
scale_y_reverse(
breaks = c(1,2,3,4,5,6,7),
label = c("Sunday", "Monday", "Tuesday", "Wednesday", "Thursday", "Friday", "Saturday")
) +
labs(title = "Weekly max HeartRate Heatmap",
caption = '@taraskaduk | taraskaduk.com',x="Time",y="") +
coord_equal() + facet_wrap(.~month,ncol=3)
## `summarise()` regrouping output by 'month', 'wday' (override with `.groups` argument)
Monthly Mean Heart rate , everyday calendar
df3 <- df %>%
dplyr::filter(type == 'HeartRate') %>%
dplyr::filter(year==2020) %>%
group_by(date,hour,minute) %>%
summarize(heartrate=mean(value)) %>%
group_by(date,hour) %>% #View()
summarize(mean.heartrate=mean(heartrate)) %>%
arrange(desc(mean.heartrate)) %>%
mutate(yday=yday(date),
wday=wday(date),
mday=mday(date),
month=month(date),
day=day(date)) %>%
group_by(date,month,mday,hour) %>%
arrange(desc(mean.heartrate))
## `summarise()` regrouping output by 'date', 'hour' (override with `.groups` argument)
## `summarise()` regrouping output by 'date' (override with `.groups` argument)
pHRmonth <- ggplot(df3,aes(x=hour, y=mday, fill=mean.heartrate)) +
geom_tile(col = 'grey40') +
scale_fill_continuous(labels = scales::comma, low = 'grey95', high = '#008FD5') +
theme(panel.grid.major = element_blank(),axis.text.x=element_text(angle=90)) +
scale_x_continuous(
breaks = c(0, 6, 12, 18),
label = c("Midnight", "6 AM", "Midday", "6 PM")
) +
scale_y_reverse(
breaks = 1:31,label=c(1," ",3," ",5," ",7," ",9," ",11," ",13," ",15," ",17," ",19," ",21," ",23," ",25," ",27," ",29," ",31)
# label = c("Sunday", "Monday", "Tuesday", "Wednesday", "Thursday", "Friday", "Saturday")
) +
labs(title = "Monthly mean heartrate Heatmap",
# caption = '@taraskaduk | taraskaduk.com',
x="Time",y="") +
coord_equal() + facet_wrap(.~month,ncol=3)
pHRmonth

Make notes on graph by geom_text()
- To writes texts on specific place in a graph, use separate data for the text info.
# data frame for text
df3.text <- tibble(month=c(3,5,9,10),mday=c(19,20,15,6),hour=12,mean.heartrate=100,label=c("Shelter in place","Synchro practice restart","Synchro fall training starts","Synchro team practice starts"))
pHRmonth + geom_text(data = df3.text,mapping=aes(label = label),size=3)



